🧮 Algorithm Title
+ + +
+🎯 Objective
+ +-
+
- Example: "This is a K-Nearest Neighbors (KNN) classifier algorithm used for classifying data points based on their proximity to other points in the dataset." +
📚 Prerequisites
+ + +-
+
- Linear Algebra Basics +
- Probability and Statistics +
- Libraries: NumPy, TensorFlow, PyTorch (as applicable) +
+
🧩 Inputs
+ +-
+
- Example: The input dataset should be in CSV format with features and labels for supervised learning algorithms. +
📤 Outputs
+ +-
+
- Example: The algorithm returns a predicted class label or a regression value for each input sample. +
+
🏛️ Algorithm Architecture
+ +-
+
- Example: "The neural network consists of 3 layers: an input layer, one hidden layer with 128 units, and an output layer with 10 units for classification." +
🏋️♂️ Training Process
+ +-
+
- Example:
-
+
- The model is trained using the gradient descent optimizer. +
- Learning rate: 0.01 +
- Batch size: 32 +
- Number of epochs: 50 +
- Validation set: 20% of the training data +
+
📊 Evaluation Metrics
+ +-
+
- Example: "Accuracy and F1-Score are used to evaluate the classification performance of the model. Cross-validation is used to reduce overfitting." +
+
💻 Code Implementation
+ +# Example: Bayesian Regression implementation
+
+import numpy as np
+from sklearn.linear_model import BayesianRidge
+import matplotlib.pyplot as plt
+
+# Generate Synthetic Data
+np.random.seed(42)
+X = np.random.rand(20, 1) * 10
+y = 3 * X.squeeze() + np.random.randn(20) * 2
+
+# Initialize and Train Bayesian Ridge Regression
+model = BayesianRidge(alpha_1=1e-6, lambda_1=1e-6, compute_score=True)
+model.fit(X, y)
+
+# Make Predictions
+X_test = np.linspace(0, 10, 100).reshape(-1, 1)
+y_pred, y_std = model.predict(X_test, return_std=True)
+
+# Display Results
+print("Coefficients:", model.coef_)
+print("Intercept:", model.intercept_)
+
+# Visualization
+plt.figure(figsize=(8, 5))
+plt.scatter(X, y, color="blue", label="Training Data")
+plt.plot(X_test, y_pred, color="red", label="Mean Prediction")
+plt.fill_between(
+ X_test.squeeze(),
+ y_pred - y_std,
+ y_pred + y_std,
+ color="orange",
+ alpha=0.3,
+ label="Predictive Uncertainty",
+)
+plt.title("Bayesian Regression with Predictive Uncertainty")
+plt.xlabel("X")
+plt.ylabel("y")
+plt.legend()
+plt.show()
+🔍 Scratch Code Explanation
+ + +Bayesian Regression is a probabilistic approach to linear regression that incorporates prior beliefs and updates these beliefs based on observed data to form posterior distributions of the model parameters. Below is a breakdown of the implementation, structured for clarity and understanding.
++
1. Class Constructor: Initialization
+class BayesianRegression:
+ def __init__(self, alpha=1, beta=1):
+ """
+ Constructor for the BayesianRegression class.
+
+ Parameters:
+ - alpha: Prior precision (controls the weight of the prior belief).
+ - beta: Noise precision (inverse of noise variance in the data).
+ """
+ self.alpha = alpha
+ self.beta = beta
+ self.w_mean = None
+ self.w_precision = None
+-
+
- Key Idea
-
+
- The
alpha(Prior precision, representing our belief in the model parameters' variability) andbeta(Precision of the noise in the data) hyperparameters are crucial to controlling the Bayesian framework. A higheralphameans stronger prior belief in smaller weights, whilebetacontrols the confidence in the noise level of the observations.
+ w_mean- Posterior mean of weights (initialized as None)
+w_precision- Posterior precision matrix (initialized as None)
+
+ - The
+
2. Fitting the Model: Bayesian Learning
+def fit(self, X, y):
+ """
+ Fit the Bayesian Regression model to the input data.
+
+ Parameters:
+ - X: Input features (numpy array of shape [n_samples, n_features]).
+ - y: Target values (numpy array of shape [n_samples]).
+ """
+ # Add a bias term to X for intercept handling.
+ X = np.c_[np.ones(X.shape[0]), X]
+
+ # Compute the posterior precision matrix.
+ self.w_precision = (
+ self.alpha * np.eye(X.shape[1]) # Prior contribution.
+ + self.beta * X.T @ X # Data contribution.
+ )
+
+ # Compute the posterior mean of the weights.
+ self.w_mean = np.linalg.solve(self.w_precision, self.beta * X.T @ y)
+Key Steps in the Fitting Process
+-
+
- Add Bias Term: The bias term (column of ones) is added to
Xto account for the intercept in the linear model.
+ -
+
Posterior Precision Matrix: + $$ + \mathbf{S}_w^{-1} = \alpha \mathbf{I} + \beta \mathbf{X}^\top \mathbf{X} + $$
+-
+
- The prior contributes \(\alpha \mathbf{I}\), which regularizes the weights. +
- The likelihood contributes \(\beta \mathbf{X}^\top \mathbf{X}\), based on the observed data. +
+ -
+
Posterior Mean of Weights: + $$ + \mathbf{m}_w = \mathbf{S}_w \beta \mathbf{X}^\top \mathbf{y} + $$
+-
+
- This reflects the most probable weights under the posterior distribution, balancing prior beliefs and observed data. +
+
+
3. Making Predictions: Posterior Inference
+def predict(self, X):
+ """
+ Make predictions on new data.
+
+ Parameters:
+ - X: Input features for prediction (numpy array of shape [n_samples, n_features]).
+
+ Returns:
+ - Predicted values (numpy array of shape [n_samples]).
+ """
+ # Add a bias term to X for intercept handling.
+ X = np.c_[np.ones(X.shape[0]), X]
+
+ # Compute the mean of the predictions using the posterior mean of weights.
+ y_pred = X @ self.w_mean
+
+ return y_pred
+Key Prediction Details
+-
+
- Adding Bias Term: The bias term ensures that predictions account for the intercept term in the model. +
- Posterior Predictive Mean:
+ $$
+ \hat{\mathbf{y}} = \mathbf{X} \mathbf{m}_w
+ $$
-
+
- This computes the expected value of the targets using the posterior mean of the weights. +
+
+
4. Code Walkthrough
+-
+
- Posterior Precision Matrix (\(\mathbf{S}_w^{-1}\)): Balances the prior (\(\alpha \mathbf{I}\)) and the data (\(\beta \mathbf{X}^\top \mathbf{X}\)) to regularize and incorporate observed evidence. +
- Posterior Mean (\(\mathbf{m}_w\)): Encodes the most likely parameter values given the data and prior. +
- Prediction (\(\hat{\mathbf{y}}\)): Uses the posterior mean to infer new outputs, accounting for both prior knowledge and learned data trends. +
+
🛠️ Example Usage
+ + +# Example Data
+X = np.array([[1.0], [2.0], [3.0]]) # Features
+y = np.array([2.0, 4.0, 6.0]) # Targets
+
+# Initialize and Train Model
+model = BayesianRegression(alpha=1.0, beta=1.0)
+model.fit(X, y)
+
+# Predict on New Data
+X_new = np.array([[4.0], [5.0]])
+y_pred = model.predict(X_new)
+
+print(f"Predictions: {y_pred}")
+-
+
- Explanation
-
+
- A small dataset is provided where the relationship between \(X\) and \(y\) is linear. +
- The model fits this data by learning posterior distributions of the weights. +
- Predictions are made for new inputs using the learned posterior mean. +
+
+
🌟 Advantages
+-
+
- Encodes uncertainty explicitly, providing confidence intervals for predictions. +
- Regularization is naturally incorporated through prior distributions. +
- Handles small datasets effectively by leveraging prior knowledge. +
⚠️ Limitations
+-
+
- Computationally intensive for high-dimensional data due to matrix inversions. +
- Sensitive to prior hyperparameters (\(\alpha, \beta\)). +
🚀 Application
+ + +Explain your application
+Explain your application
+ +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+.jpg) +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+ 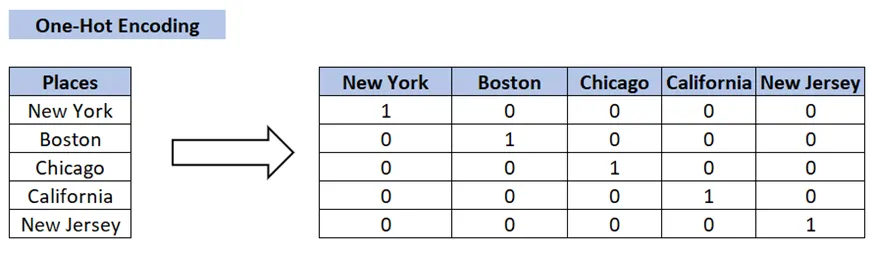 +
+ .webp) +
+ 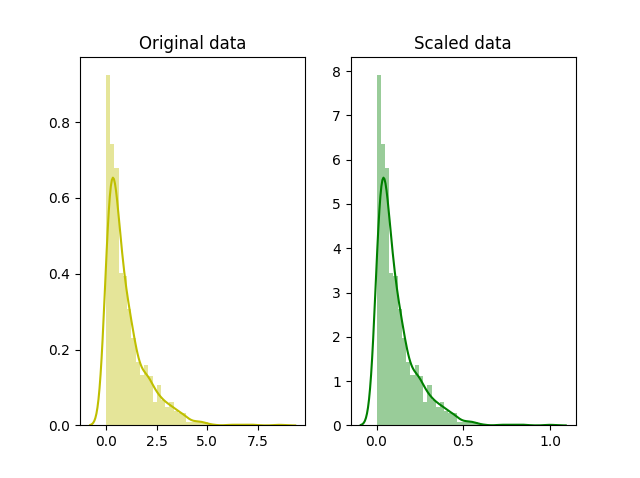 +
+  +
+ 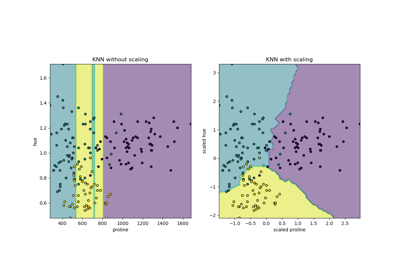 +
+  +
+  +
+  +
+ 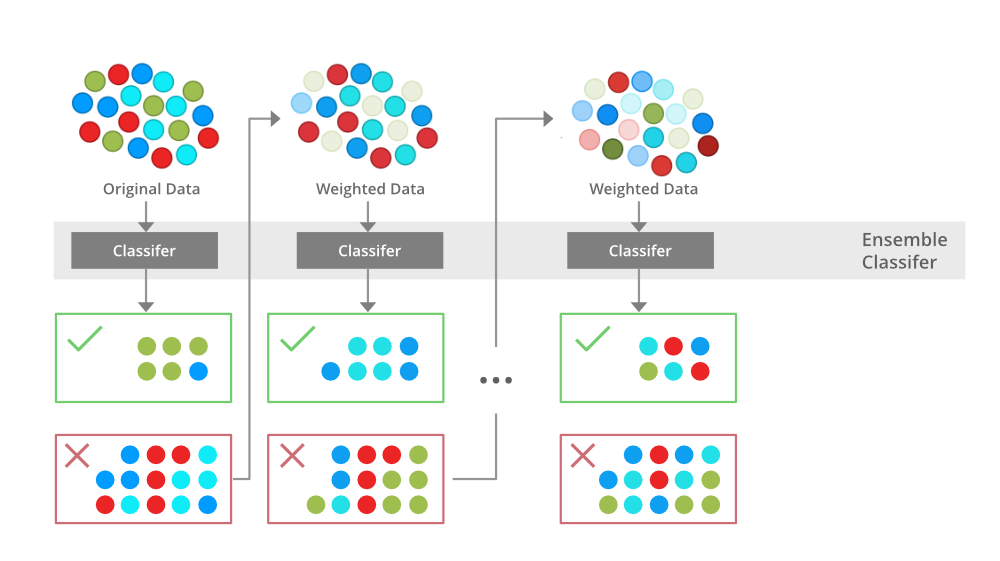 +
+  +
+  +
+  +
+  +
+  +
+  +
+ 
 +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+ :max_bytes(150000):strip_icc()/Descriptive_statistics-5c8c9cf1d14d4900a0b2c55028c15452.png) +
+  +
+  +
+  +
+