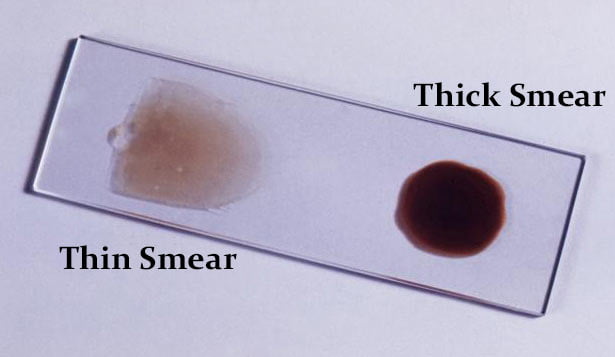
Malaria is a blood disease caused by the Plasmodium parasites transmitted through the bite of female Anopheles mosquito. Microscopists commonly examine thick and thin blood smears to diagnose disease and compute parasitemia. However, their accuracy depends on smear quality and expertise in classifying and counting parasitized and uninfected cells. Such an examination could be arduous for large-scale diagnoses resulting in poor quality. State-of-the-art image-analysis based computer- aided diagnosis (CADx) methods using machine learning (ML) techniques, applied to microscopic images of the smears using hand-engineered features demand expertise in analyzing morphological, textural, and positional variations of the region of interest (ROI). In contrast, Convolutional Neural Networks (CNN), a class of deep learning (DL) models promise highly scalable and superior results with end-to-end feature extraction and classification. Automated malaria screening using DL techniques could, therefore, serve as an effective diagnostic aid.
This repository presents a deep learning model for blood smear image analysis using a pretrained network ResNet50
- Pytorch
- CUDA
- Google Colab
- Python 3.8
- Numpy
This repo already contains the images data in the "cell_images" folder. Alternatively you can download the dataset from here: https://ceb.nlm.nih.gov/repositories/malaria-datasets/
- smear_analyser_cpu.ipynb: version of the notebook trained on CPU
- smear_analyser_gpu.ipynb: version of the notebook trained on GPU(Google Colab)
- test_image.py: python file using trained model to test images
- smear_analyser.pt: Model file