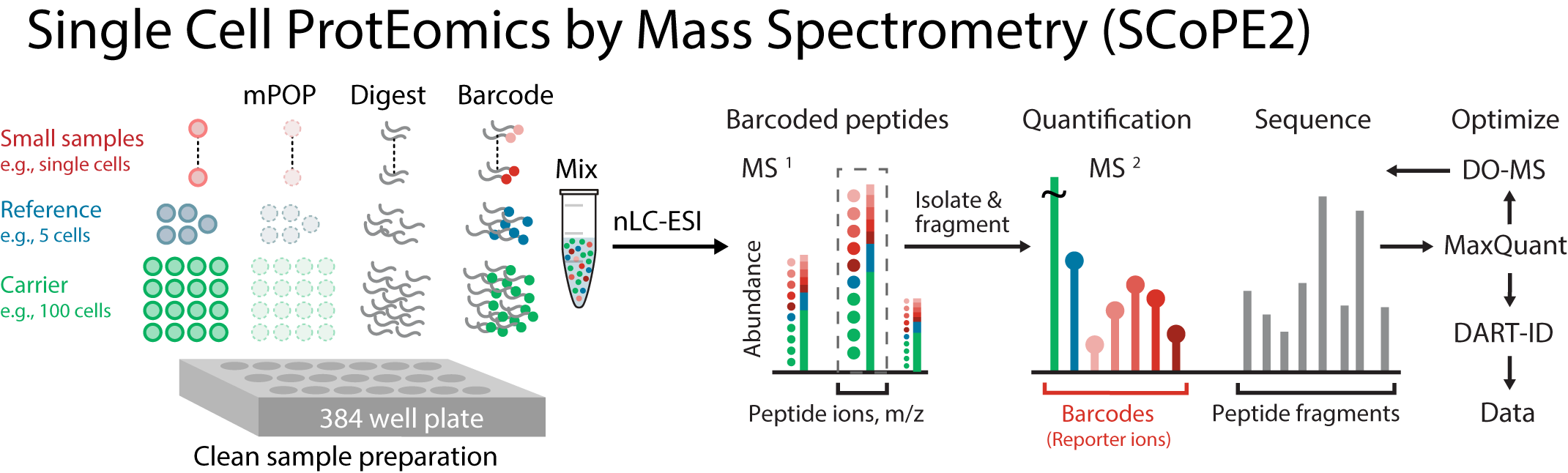
This is a second generation method building upon SCoPE-MS
This application has been tested on R >= 3.5.0, OSX 10.14 / Windows 7/8/10. R can be downloaded from the main R Project page or downloaded with the RStudio Application. All modules are maintained for MaxQuant >= 1.6.0.16.
Each thematic part of the SCoPE2_analysis.R script is bounded by an R "region". These can be minimized when not in use. You will need to create "figs","dat", and "code" sub-directories.
To execute the ladder_experiment_analysis.R script you will have to change the path to the ladder experiment evidence file provided on MassIVE, as well as install packages ggpubr and reshape2 from CRAN. The ladder_experiment_design.csv is not necessary for script execution.
The manuscript is freely available on bioRxiv: Specht et al., 2019 and at Genome Biology Specht et al., 2021.
Contact the authors by email: nslavov{at}northeastern.edu.
The SCoPE2 code is distributed by an MIT license.
Please feel free to contribute to this project by opening an issue or pull request.
For any bugs, questions, or feature requests, please use the GitHub issue system to contact the developers.