| title | tags | authors | affiliations | date | cito-bibliography | event | biohackathon_name | biohackathon_url | biohackathon_location | git_url | group | authors_short | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Secure Processing Environments as a Service in the de.NBI Cloud |
|
|
|
21 February 2025 |
paper.bib |
BHG2024 |
BioHackathon Germany 2024 |
Kassel, Germany, 2024 |
Project 3 |
Nils Hoffmann, Sven Twardziok \emph{et al.} |
In biomedical research, sensitive data from humans is a critical asset for the ability to carry out essential research in even potentially critical situations, e.g. as proven during the COVID-19 pandemic. Providing easy access to data speeds up the research process, resulting in faster development of new drugs, or exploring and further understanding of rare diseases. With the German model project for comprehensive diagnostics and therapy identification using genome sequencing for rare and oncological diseases according to §64e, Sozialgesetzbuch and the European Health Data Space (EHDS), more clinical data from daily routine will be available for translational research in the future in Germany and in Europe. However, a high level of protection of sensitive data must be implemented. Today, different approaches for implementing secure environments exist. The concept of the 5 safes (Safe projects, Safe people, Safe settings, Safe data, Safe outputs) is used to build Trusted Research Environments (TRE) in the UK and the EHDS calls for the development of Secure Processing Environments (SPE) for the processing of health data for research. In both approaches, technical solutions are used to ensure that sensitive data is highly protected.
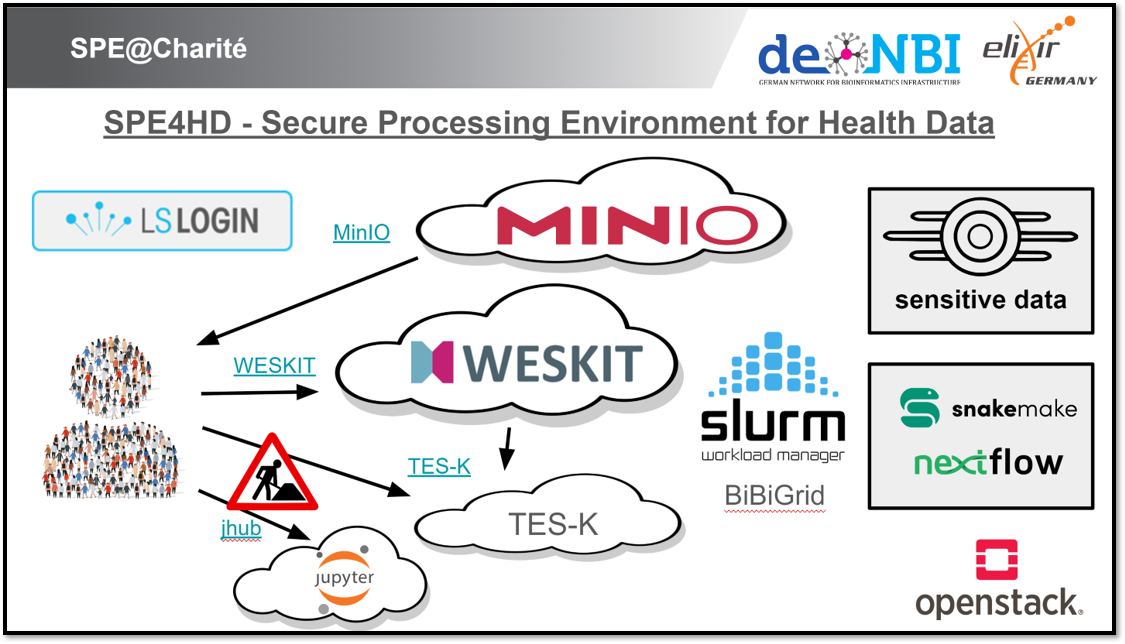
In this Biohackathon Germany project, we used existing services from ELIXIR Europe [@ELIXIR_2021] as well as external tools to create a technical foundation for reusable Secure Processing Environments in the de.NBI cloud as well as in the SURF research cloud. Specifically, the authentication was based on Life Science Login for authentication and authorization. We established the open source object storage platform MinIO for sharing sensitive data with users. Hereby, we extended deployment of virtual machines with MinIO access, so that users are able to access shared data in their custom environments. For execution of workflows on protected data, we developed a demonstration platform combining the workflow execution system WESkit with the BiBiGrid system for creation of a SLURM cluster. The TES-K software has been deployed to multiple cloud locations to connect these sites to a federated processing network operated by ELIXIR.
MinIO is an open source object storage platform that can be used for sharing data with users. Regarding sharing sensitive data, it is required to authenticate users and make sure that only a authorized users will have access to the sensitive data. We connected MinIO instances in the de.NBI cloud and in the SURF cloud to LS-LOGIN for authentication of users. The instruction on how to integrate LS-LOG in MinIO was submitted to the Cloud&AAI documentation during the BioHackathon. LS-Login can be activated in MinIO either by using the MinIO console using the OIDC configuration or by setting environmental variables (MinIO OIDC Documentation).
- Config URL (
MINIO_IDENTITY_OPENID_CONFIG_URL) - Client ID (
MINIO_IDENTITY_OPENID_CLIENT_ID)- Id of the LS-Login service
- Client secret (
MINIO_IDENTITY_OPENID_CLIENT_SECRET)- Secret of the LS-Login service
- Display Name (
MINIO_IDENTITY_OPENID_DISPLAY_NAME)- A human readable label for the login button (e.g.
LS-Login)
- A human readable label for the login button (e.g.
- Scopes (
MINIO_IDENTITY_OPENID_SCOPES)- Scopes that will be requested from LS-Login (e.g.
openid,email,profile)
- Scopes that will be requested from LS-Login (e.g.
- Role policy (
MINIO_IDENTITY_OPENID_ROLE_POLICY)- Name of a policy in MinIO that will be used to manage access of LS-Login users (e.g.
readonly).
- Name of a policy in MinIO that will be used to manage access of LS-Login users (e.g.
- Claim User Info (
MINIO_IDENTITY_OPENID_CLAIM_USERINFO)- Allow MinIO to request the userinfo endpoint for additional information (
on).
- Allow MinIO to request the userinfo endpoint for additional information (
MinIO supports two different mechanisms for authorization of users with OIDC (MinIO OIDC authorization). It is recommended to use the RolePolicy flow. Here, all LS-Login users in MinIO will be assigned to one or more policies. These policies can control access to specific buckets by group membership; e.g. require that users belong to a specific LS-AAI group (see policy based access control).
In order to mount MinIO to a virtual machine we modified an existing Ansible playbook that mounts WebDav connections. The new Ansible playbook mounts a specified MinIO S3-based connection. It uses RClone to establish the connection and a systemd mount to mount the MinIO resource to the local filesystem. Whichever policies and restrictions apply for the created set of MinIO credentials, will of course also apply to the established connection. The Ansible playbook is written to be a SURF Research Cloud catalog item component. However, the playbook has only a few little twists that would be specific to SURF Research Cloud. If you interpolate a little with the four "{{ variable_names }}", you can modify it for whatever your purpose is. The four variables are the access key id, and the corresponding access key secret. They have been created in MinIO, before. In Research Cloud, they are kept in the secrets vault of the collaboration (=user project). The third variable is simply the URL of the MinIO API. A fourth parameter is the directory name that is chosen for the mount. It defaults to "minio". The entire path being "~/data/<chosen dirname>". The last two are passed as parameters from the catalog item, in Research Cloud. This component can be applied to any Ubuntu VM. It may run on other distributions with little or no modification.
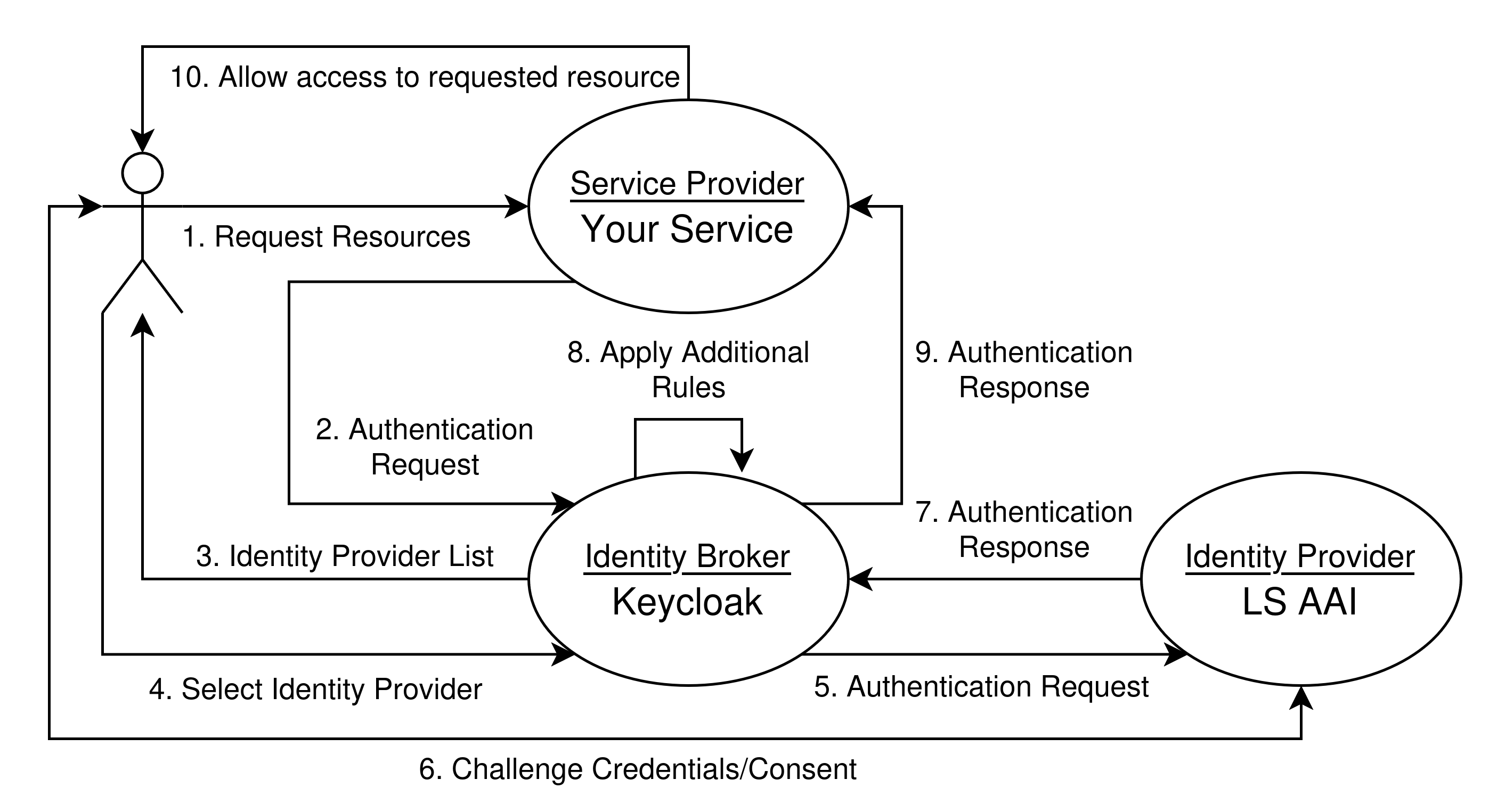
In this section we explain how to add a Keycloak instance between your service (e.g. MinIO) and your identity provider (e.g. LS AAI). This setup allows for a more direct management of groups and rights since you are able to handle those on the keycloak level instead of the individual identity provider level.
Users 1. request a resource from the service in question which 2. requests authentication from the identity broker Keycloak. Users then get the option to 3. identify via their chosen Identity Provider (e.g. LS AAI). Users 4. select an option - for now we assume LS AAI. 5. Keycloak asks LS AAI to handle authentication. 6. LS AAI will now ask users to login and forwards that response to Keycloak. Keycloak 8. might apply additional rules 9. then forwards its own authentication response and finally the service 10. your service allows access or denies it.
Instead of a single authentication request and response, this setup requires two. One request by the service directed at Keycloak which triggers the second request and response pair: a request by Keycloak to the final identity provider and one response from that identity provider to Keycloak - and then Keycloak forwards its own response back to your service.
Technical documentation is available in the GitHub repository.
TES-K was deployed at different sites according to the online tutorial provided in the ELIXIR-Cloud-AAI documentation.
-
On SURF Research Cloud: There is a catalog item (=VM template) on the Dutch Research Cloud that creates a TES-K cluster in a Virtual Machine, with a few clicks. It can be addressed from external. So we still would have to add authentication or at least constrain the allowed ip-ranges. Also, adding S3 storage may make it more useful.
-
Bielefeld: TES-K was set up using custom Terraform and Ansible scripts together with customized Helm Charts to deploy a customized Kubermatic KubeOne Kubernetes cluster and TES-K.
-
At the Charité site used an existing OpenStack project and deployed TES-K in a connected Kubermatic cluster to make the tool available at the URL https://spe4hd.tesk.bihealth.org. The OpenStack setup with Kubermatic required to create a custom
LoadBalancerservice in the defined namespace usingkubectlto connect the public IP address with the TES-K application.
At the de.NBI cloud site at the Charité we established a demonstration platform during the BioHackathon in order to execute demonstration workflows in a controlled environment. The platform is based on an instance of the workflow execution service WESkit, which submits workflows to a SLURM cluster that was build in the cloud using the tool BiBiGrid. Authenticated (LS Login) and authorized users are able to execute secure and validated Snakemake and Nextflow workflows on sensitive data using WESkit, an implementation of the GA4GH WES standard. The actual computation will be relayed to a SLURM cluster system based on BiBiGrid that is deployed within the secure processing environment. WESkit will execute the workflows on behalf of the users on the SLURM cluster. The sensitive data are available via the cluster's file system for authorized users. This allows certain data sets to be shared, while other data remains protected from user access. Scrips and documentation are available in the GitHub repository.
In this BioHackathon Germany project, we utilized various open-source services to facilitate the handling of sensitive data in the de.NBI Cloud and the SURF Cloud. Building on previous BioHackathon Europe projects (2021, 2022, and 2024), this initiative aligns with the activities of the ELIXIR Compute Platform. The project's outcomes support scientists in establishing secure processing environments and the development of project-specific cloud platforms [@f1000cloud_2024]. Additionally, through the extension of the ELIXIR Cloud AAI documentation, the results are available to users within the ELIXIR community.
Using the tools WESkit and BiBiGrid we developed a demonstration platform for processing sensitive data in the cloud. Workflow systems generally support the implementation of FAIR principles [@FAIR_2016] and can contribute to the secure processing of sensitive data. The GA4GH defines various specifications for executing workflows in the cloud through its Cloud Work Stream [@GA4GH_2021]. WESkit implements GA4GH WES and supports workflow systems Snakemake [@Nextflow_2017] and Nextflow [@Snakemake_2021]. By allowing only secure workflows, Weskit ensures secure processing of sensitive data. BiBiGrid was used for setting up cluster systems and using the clusters permission systems to ensure users can only access files they are authorized for. This combination of WESkit and BiBiGrid can advance the establishment of Secure Processing Environments (SPEs) specifically in the de.NBI Cloud.
- Main Hackathon results repository: https://github.com/deNBI/2024_BioHackathon_DE_SPE
- BiBiGrid SLURM cluster repository: https://github.com/BiBiServ/bibigrid
- SURF Minio S3 examples repository: https://gitlab.com/rsc-surf-nl/plugins/mount-minio-s3
We acknowledge support by de.NBI - the German Network for Bioinformatics Infrastructure and the de.NBI BioHackathon Germany 2024.