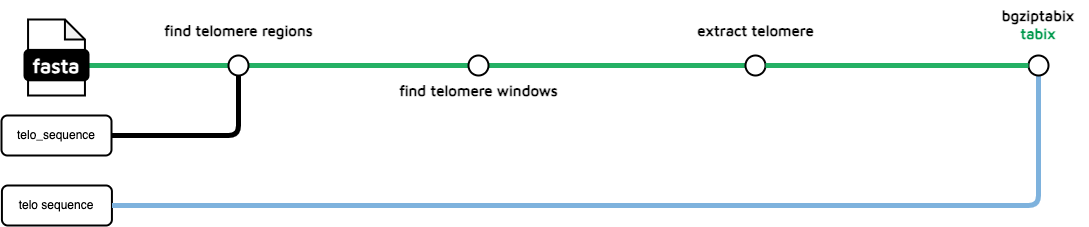
Seqtk-telo from Heng Li has long been incorporated into TreeValGal now, making this complicated TreeVal workflow trivial to implement in Galaxy.
It takes the fasta being reported, an optional repeat and the bed goes straight to the JBrowse2 tool for tabix indexing and display. It is the tool at the top of the second column from the right of this part of the TreeValGal workflow. Two inputs and a single tool is about as simple as it gets.
Compare this to the two Java binaries of unknown provenance used in TreeVal as described below. No provenance or references are provided so it's hard to know what the Sanger version really does. Might be much better, but this is risibly simple in comparison. Choice is between Heng Li and unknown Java executables. Not hard to decide.
No Galaxy system administrator would be thrilled at the prospect of running what NextFlow developers regard as state of the art security.
According to Sanger's documentation, TreeVal represents "best practice".
seqtk-telo is now available as an IUC tool in the toolshed and may provide a basis for a replacement after evaluation and advice from VGP.
Binary and jar files make this a good candidate for a rewrite using existing Galaxy tools with executables of known provenance. It seems to write masked telomere windows into a tabix file and that might already be part of a VGP workflow?
In particular, this might be the origin of the NF /tree/bin telomere jar and binary? There's a build.sh for a jar and some c code to search for a user supplied or vertebrate default sequence. If that's true, getting those dependencies into Conda becomes feasible and new Galaxy tools are then possible for a new workflow.
As a prototype, a temporary tool with using the existing executable, jar files and shell scripts would be useful as a demonstration and test for consistent output with TreeVal test data, when a sustainable long term solution is built for the final production version. Potential tools that we could use include:
The DDL has function calls explained below. Most of the rest of the 76 line DDL file is not going to be needed other than to figure out exactly how each function gets parameters supplied to the actual command lines.
#!/usr/bin/env nextflow
//
// MODULE IMPORT BLOCK
//
include { FIND_TELOMERE_REGIONS } from '../../modules/local/find_telomere_regions'
include { FIND_TELOMERE_WINDOWS } from '../../modules/local/find_telomere_windows'
include { EXTRACT_TELO } from '../../modules/local/extract_telo'
include { TABIX_BGZIPTABIX } from '../../modules/nf-core/tabix/bgziptabix'
workflow TELO_FINDER {
take:
max_scaff_size // Channel: val(size of largest scaffold in bp)
reference_tuple // Channel: tuple [ val(meta), path(fasta) ]
teloseq
main:
ch_versions = Channel.empty()
//
// MODULE: FINDS THE TELOMERIC SEQEUNCE IN REFERENCE
//
FIND_TELOMERE_REGIONS (
reference_tuple,
teloseq
)
ch_versions = ch_versions.mix( FIND_TELOMERE_REGIONS.out.versions )
//
// MODULE: GENERATES A WINDOWS FILE FROM THE ABOVE
//
FIND_TELOMERE_WINDOWS (
FIND_TELOMERE_REGIONS.out.telomere
)
ch_versions = ch_versions.mix( FIND_TELOMERE_WINDOWS.out.versions )
//
// MODULE: EXTRACTS THE LOCATION OF TELOMERIC SEQUENCE BASED ON THE WINDOWS
//
EXTRACT_TELO (
FIND_TELOMERE_WINDOWS.out.windows
)
ch_versions = ch_versions.mix( EXTRACT_TELO.out.versions )
//
// LOGIC: Adding the largest scaffold size to the meta data so it can be used in the modules.config
//
EXTRACT_TELO.out.bed
.combine(max_scaff_size)
.map {meta, row, scaff ->
tuple(
[ id : meta.id,
max_scaff : scaff >= 500000000 ? 'csi': '' ],
file( row )
)
}
.set { modified_bed_ch }
//
// MODULE: BGZIP AND TABIX THE OUTPUT FILE
//
TABIX_BGZIPTABIX (
modified_bed_ch
)
ch_versions = ch_versions.mix( TABIX_BGZIPTABIX.out.versions )
emit:
bed_file = EXTRACT_TELO.out.bed
bed_gz_tbi = TABIX_BGZIPTABIX.out.gz_tbi
bedgraph_file = EXTRACT_TELO.out.bedgraph
versions = ch_versions.ifEmpty(null)
}
executes a binary in the /tree/bin directory (!)
find_telomere ${file} $telomereseq > ${prefix}.telomere
That binary is interesting but of unknown origin:
>file find_telomere
find_telomere: ELF 64-bit LSB pie executable, x86-64, version 1 (SYSV), dynamically linked, interpreter /lib64/ld-linux-x86-64.so.2, for GNU/Linux 3.2.0, BuildID[sha1]=c513d67e8fd6d09eed4e44c095c35ed40e0144cd, not stripped
executes a java jar in the /tree/bin directory
java ${telomere_jvm_params} -cp ${projectDir}/bin/${telomere_jar} \ FindTelomereWindows $file $telomere_window_cut > ${prefix}.windows
Again, if this whole thing can be replaced with something more sustainable than these binaries and jar files?
runs awk and sed to write bed and bedgraphs
$/
cat "${file}" |awk '{print $2"\t"$4"\t"$5}'|sed 's/>//g' > ${prefix}_telomere.bed
cat "${file}" |awk '{print $2"\t"$4"\t"$5"\t"$6}'|sed 's/>//g' > ${prefix}_telomere.bedgraph
so again can be replaced with something simpler.
TABIX_BGZIPTABIX runs some shell lines like the tabix in gap_finder:
bgzip --threads ${task.cpus} -c $args $input > ${prefix}.${input.getExtension()}.gz
tabix $args2 ${prefix}.${input.getExtension()}.gz