-
Notifications
You must be signed in to change notification settings - Fork 1
/
Copy pathlesson_06_pymc3-bayes.py
800 lines (547 loc) · 23.5 KB
/
lesson_06_pymc3-bayes.py
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
398
399
400
401
402
403
404
405
406
407
408
409
410
411
412
413
414
415
416
417
418
419
420
421
422
423
424
425
426
427
428
429
430
431
432
433
434
435
436
437
438
439
440
441
442
443
444
445
446
447
448
449
450
451
452
453
454
455
456
457
458
459
460
461
462
463
464
465
466
467
468
469
470
471
472
473
474
475
476
477
478
479
480
481
482
483
484
485
486
487
488
489
490
491
492
493
494
495
496
497
498
499
500
501
502
503
504
505
506
507
508
509
510
511
512
513
514
515
516
517
518
519
520
521
522
523
524
525
526
527
528
529
530
531
532
533
534
535
536
537
538
539
540
541
542
543
544
545
546
547
548
549
550
551
552
553
554
555
556
557
558
559
560
561
562
563
564
565
566
567
568
569
570
571
572
573
574
575
576
577
578
579
580
581
582
583
584
585
586
587
588
589
590
591
592
593
594
595
596
597
598
599
600
601
602
603
604
605
606
607
608
609
610
611
612
613
614
615
616
617
618
619
620
621
622
623
624
625
626
627
628
629
630
631
632
633
634
635
636
637
638
639
640
641
642
643
644
645
646
647
648
649
650
651
652
653
654
655
656
657
658
659
660
661
662
663
664
665
666
667
668
669
670
671
672
673
674
675
676
677
678
679
680
681
682
683
684
685
686
687
688
689
690
691
692
693
694
695
696
697
698
699
700
701
702
703
704
705
706
707
708
709
710
711
712
713
714
715
716
717
718
719
720
721
722
723
724
725
726
727
728
729
730
731
732
733
734
735
736
737
738
739
740
741
742
743
744
745
746
747
748
749
750
751
752
753
754
755
756
757
758
759
760
761
762
763
764
765
766
767
768
769
770
771
772
773
774
775
776
777
778
779
780
781
782
783
784
785
786
787
788
789
790
791
792
793
794
795
796
797
798
799
800
# coding: utf-8
# 我想医生们也还是需要大量进行"传统"的统计学分析的. 这几天正好看到了一个关于贝叶斯分析的PyMC包, 用来做概率计算. 下面的文章里讲解了临床最常见的一些统计应用, 比如参数估计, 组间比较. 看着写起来也不困难, 供参考.
#
# ----
#
# 本文翻译自[Eric Ma](https://github.com/ericmjl)在PyCon2017上的演讲Bayesian Statistical Analysis with Python
#
# 讲解了使用PyMC3进行基本的贝叶斯统计分析过程. 是很好的PyMC3和贝叶斯统计分析的入门教程.
#
# * [演讲Slides原文](https://github.com/ericmjl/bayesian-stats-talk)
# * [演讲视频(youtube)](https://www.youtube.com/watch?time_continue=3&v=p1IB4zWq9C8)
# In[1]:
# Imports
import pymc3 as pm # python的概率编程包
import numpy.random as npr # numpy是用来做科学计算的
import numpy as np
import matplotlib.pyplot as plt # matplotlib是用来画图的
import matplotlib as mpl
from collections import Counter # ?
import seaborn as sns # ?
# import missingno as msno # 用来应对缺失的数据
# Set plotting style
# plt.style.use('fivethirtyeight')
sns.set_style('white')
sns.set_context('poster')
get_ipython().magic('load_ext autoreload')
get_ipython().magic('autoreload 2')
get_ipython().magic('matplotlib inline')
get_ipython().magic("config InlineBackend.figure_format = 'retina'")
import warnings
warnings.filterwarnings('ignore')
# # 使用python进行贝叶斯统计分析
#
# Eric J. Ma, MIT Biological Engineering, Insight Health Data Science Fellow, NIBR Data Science
#
# PyCon 2017, Portland, OR; PyData Boston 2017, Boston, MA
#
# - HTML Notebook on GitHub: [**ericmjl**.github.io/**bayesian-stats-talk**](https://ericmjl.github.io/bayesian-stats-talk)
# - Twitter: [@ericmjl](https://twitter.com/ericmjl)
# ## 本讲座优点
# - **最少的专业术语:**让我们专注于分析原理,而不是术语。 *例如。 将不会解释A / B测试,spike & slab回归,共轭分布... *
# - **帕雷托原则:**你需要的80%都是基本知识
# - **享受讲座:**专注于贝叶斯,以后再看代码!
# ## 假设已经掌握的知识
# - 熟悉Python:
# - 对象 & 方法
# - context manager syntax
# - 了解基本的统计学术语:
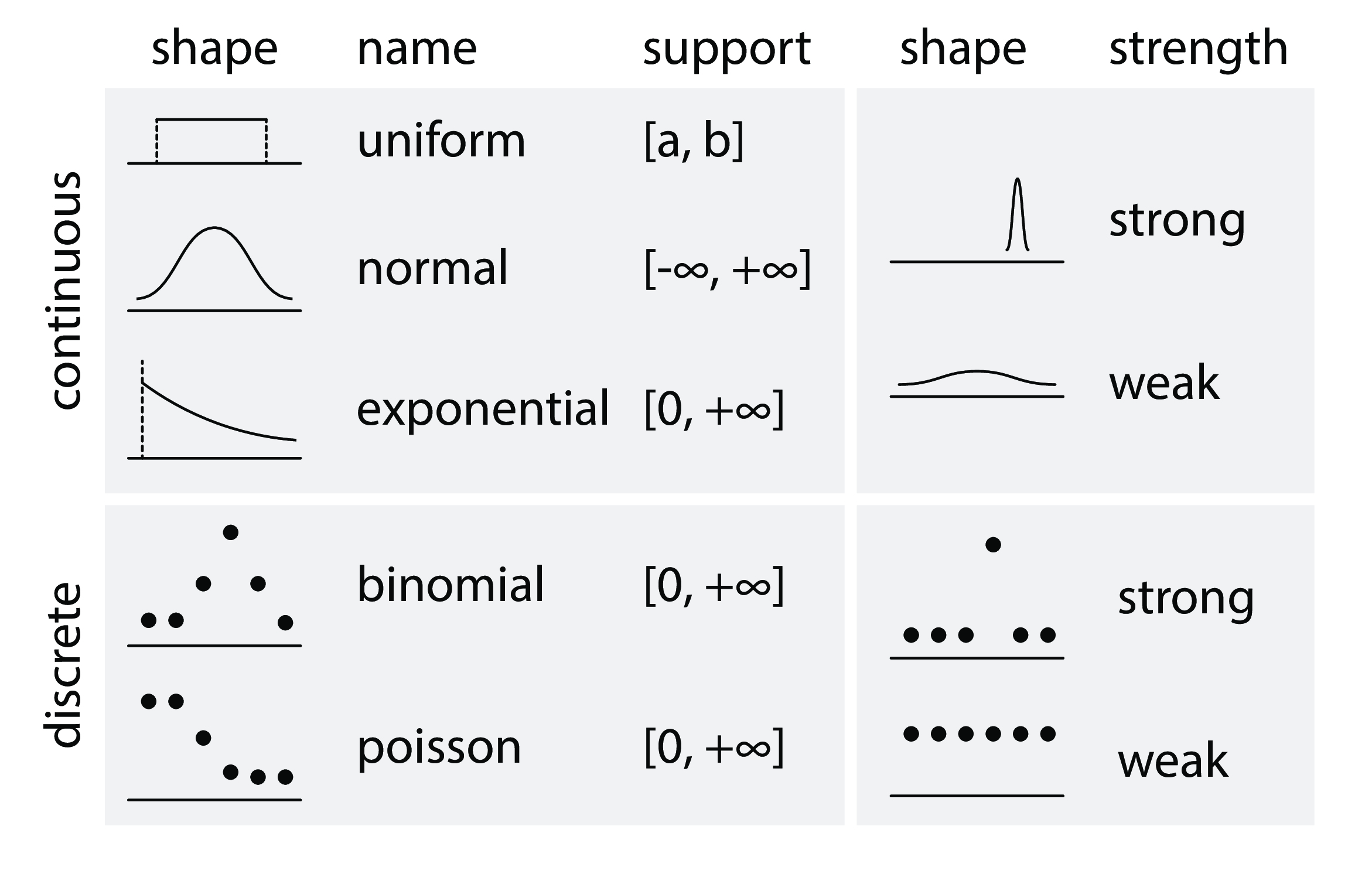
# - mean: 均值
# - variance: 方差
# - interval: 置信区间
# 
# ## 贝叶斯公式
# $$ P(H|D) = \frac{P(D|H)P(H)}{P(D)} $$
#
# - $ P(H|D)$:在给定数据的情况下, 假设为真的概率。(Probability that the hypothesis is true given the data.)
# - $ P(D|H)$:假设为真时, 数据事件发生的概率。(Probability of the data arising given the hypothesis.)
# - $ P(H)$:假设发生的概率。(Probability that the hypothesis is true, globally.)
# - $ P(D)$:数据事件发生的概率。(Probability of the data arising, globally.)
#
# 
# ## 贝叶斯主义者的思维方式
#
# > 根据证据不断更新信念
# ## `pymc3`
#
# 
#
# - 用于指定统计模型的**统计分布**,**抽样算法**和**语法**的库
# - 一切都用Python!
# # 常见的统计分析问题
# - **参数估计**: "真实值是否等于X"
#
#
# - **比较两组实验数据**: "实验组是否与对照组不同? "
# # 问题1: 参数估计
#
# "真实值是否等于X?"
#
# 或者说
#
# "给定数据,对于感兴趣的参数,可能值的概率分布是多少?"
# # 例 1: 抛硬币问题
#
# 我把我的硬币抛了$ n $次,正面是$ h $次。 这枚硬币是有偏心的吗?
# ## 参数估计问题parameterized problem
#
# “我想知道的是抛出正面的概率$ p $,假设$ n $次抛硬币中有$ h $次观察到是正面,$ p $的值是否足够接近$ 0.5 $,比如在$ [0.48,0.52] $?“
# ## 先验假设
#
# - 对参数预先的假设分布: $ p \sim Uniform(0, 1) $
# - likelihood function(似然函数, 翻译这词还不如英文原文呢): $ data \sim Bernoulli(p) $
# 
# In[2]:
# 产生所需要的数据
from random import shuffle
total = 30
n_heads = 11
n_tails = total - n_heads
tosses = [1] * n_heads + [0] * n_tails
shuffle(tosses)
# ## 数据
# In[3]:
print(tosses)
# In[4]:
def plot_coins():
fig = plt.figure()
ax = fig.add_subplot(1,1,1)
ax.bar(list(Counter(tosses).keys()), list(Counter(tosses).values()))
ax.set_xticks([0, 1])
ax.set_xticklabels(['tails', 'heads'])
ax.set_ylim(0, 20)
ax.set_yticks(np.arange(0, 21, 5))
return fig
# In[5]:
fig = plot_coins()
plt.show()
# ## 代码
# In[6]:
# Context manager syntax. `coin_model` is **just**
# a placeholder
with pm.Model() as coin_model:
# Distributions are PyMC3 objects.
# Specify prior using Uniform object.
p_prior = pm.Uniform('p', 0, 1)
# Specify likelihood using Bernoulli object.
like = pm.Bernoulli('likelihood', p=p_prior,
observed=tosses)
# "observed=data" is key
# for likelihood.
# ## MCMC Inference Button (TM)
# In[7]:
with coin_model:
# don't worry about this:
step = pm.Metropolis()
# focus on this, the Inference Button:
coin_trace = pm.sample(2000, step=step)
# ## 结果
# In[8]:
pm.traceplot(coin_trace)
plt.show()
# In[9]:
pm.plot_posterior(coin_trace[100:], color='#87ceeb',
rope=[0.48, 0.52], point_estimate='mean',
ref_val=0.5)
plt.show()
# - <font style="color:black; font-weight:bold">95% highest posterior density (HPD, 大概类似于置信区间)</font> 包含了 <font style="color:red; font-weight:bold">region of practical equivalence (ROPE, 实际等同区间)</font>.
# - 需要更多的数据!
# # 模式
#
# 1. 使用统计分布参数化您的问题
# 1. 修正你的模型结构
# 1. 在PyMC3中编写模型,点击**Inference 按钮<sup>TM</sup>**
# 1. 根据后验分布进行解释
# 1. (可选)如果有新信息,修改模型结构。
# # 例 2: 药品活性问题
#
# 我有一个新开发的分子X; X在阻止流感病毒复制方面有多好?
# <!-- mention verbally about the context: flu, replicating, need molecule to stop it -->
# ## 实验
#
# - 测试X的浓度范围, 测量流感活性
#
# - 计算 **IC<sub>50</sub>**: 能够抑制病毒复制活性50%的X浓度.
# ## data
#
# 
# In[10]:
import numpy as np
chem_data = [(0.00080, 99),
(0.00800, 91),
(0.08000, 89),
(0.40000, 89),
(0.80000, 79),
(1.60000, 61),
(4.00000, 39),
(8.00000, 25),
(80.00000, 4)]
import pandas as pd
chem_df = pd.DataFrame(chem_data)
chem_df.columns = ['concentration', 'activity']
chem_df['concentration_log'] = chem_df['concentration'].apply(lambda x:np.log10(x))
# df.set_index('concentration', inplace=True)
# ## 参数化问题parameterized problem
#
# 给定数据, 求出化学物质的**IC<sub>50</sub>**值是多少, 并且求出置信区间( 原文中the uncertainty surrounding it, 后面看类似置信区间的含义)?
# ## 先验知识
#
# - 由药学知识已知测量函数(measurement function): $ m = \frac{\beta}{1 + e^{x - IC_{50}}} $
# - 测量函数中的参数估计, 来自先验知识: $ \beta \sim HalfNormal(100^2) $
# - 关于感兴趣参数的先验知识: $ log(IC_{50}) \sim ImproperFlat $
# - likelihood function: $ data \sim N(m, 1) $
# 
# ## 数据
# In[11]:
def plot_chemical_data(log=True):
fig = plt.figure(figsize=(10,6))
ax = fig.add_subplot(1,1,1)
if log:
ax.scatter(x=chem_df['concentration_log'], y=chem_df['activity'])
ax.set_xlabel('log10(concentration (mM))', fontsize=20)
else:
ax.scatter(x=chem_df['concentration'], y=chem_df['activity'])
ax.set_xlabel('concentration (mM)', fontsize=20)
ax.set_xticklabels([int(i) for i in ax.get_xticks()], fontsize=18)
ax.set_yticklabels([int(i) for i in ax.get_yticks()], fontsize=18)
plt.hlines(y=50, xmin=min(ax.get_xlim()), xmax=max(ax.get_xlim()), linestyles='--',)
return fig
# In[12]:
fig = plot_chemical_data(log=True)
plt.show()
# ## 代码
# In[13]:
with pm.Model() as ic50_model: # 都是以这句开头, with pm.Models() as 自己取个名字:
beta = pm.HalfNormal('beta', sd=100**2) # 每个参数需要规定分布, 用pm.xxx定义了分布函数
ic50_log10 = pm.Flat('IC50_log10') # Flat prior
# MATH WITH DISTRIBUTION OBJECTS! # 测量函数的计算过程, 这个也是来自于先验知识
measurements = beta / (1 + np.exp(chem_df['concentration_log'].values -
ic50_log10))
y_like = pm.Normal('y_like', mu=measurements,
observed=chem_df['activity']) # 这是啥?
# Deterministic transformations.
ic50 = pm.Deterministic('IC50', np.power(10, ic50_log10)) # ic50_log10是在对数域, 要转换回来
# ## MCMC Inference Button (TM)
# In[14]:
with ic50_model: # 在之前定义的模型中模拟
step = pm.Metropolis() # 标准步骤, 照写
ic50_trace = pm.sample(100000, step=step) # 随机模拟过程, 重采样的次数手工指定
# In[15]:
pm.traceplot(ic50_trace[2000:], varnames=['IC50_log10', 'IC50']) # live: sample from step 2000 onwards.
plt.show()
# ## 结果
# In[16]:
pm.plot_posterior(ic50_trace[4000:], varnames=['IC50'],
color='#87ceeb', point_estimate='mean')
plt.show()
# 该化学物质的 IC<sub>50</sub> 大约在[2 mM, 2.4 mM] (95% HPD). 这不是个好的药物候选者. 在这个问提上不确定性影响不大, 看看单位数量级就知道IC<sub>50</sub>在毫摩的物质没什么用...
# # 第二类问题: 实验组之间的比较
#
# "实验组和对照组之间是否有差别? "
# # 例 1: 药品对IQ的影响问题
#
# 药品治疗是否影响(提高)IQ分数?
#
# (documented in Kruschke, 2013, example modified from PyMC3 documentation)
# In[17]:
drug = [ 99., 110., 107., 104., 103., 105., 105., 110., 99.,
109., 100., 102., 104., 104., 100., 104., 101., 104.,
101., 100., 109., 104., 105., 112., 97., 106., 103.,
101., 101., 104., 96., 102., 101., 100., 92., 108.,
97., 106., 96., 90., 109., 108., 105., 104., 110.,
92., 100.]
placebo = [ 95., 105., 103., 99., 104., 98., 103., 104., 102.,
91., 97., 101., 100., 113., 98., 102., 100., 105.,
97., 94., 104., 92., 98., 105., 106., 101., 106.,
105., 101., 105., 102., 95., 91., 99., 96., 102.,
94., 93., 99., 99., 113., 96.]
def ECDF(data):
x = np.sort(data)
y = np.cumsum(x) / np.sum(x)
return x, y
def plot_drug():
fig = plt.figure()
ax = fig.add_subplot(1,1,1)
x_drug, y_drug = ECDF(drug)
ax.plot(x_drug, y_drug, label='drug, n={0}'.format(len(drug)))
x_placebo, y_placebo = ECDF(placebo)
ax.plot(x_placebo, y_placebo, label='placebo, n={0}'.format(len(placebo)))
ax.legend()
ax.set_xlabel('IQ Score')
ax.set_ylabel('Cumulative Frequency')
ax.hlines(0.5, ax.get_xlim()[0], ax.get_xlim()[1], linestyle='--')
return fig
# In[18]:
# Eric Ma自己很好奇, 从频率主义的观点, 差别是否已经是具有"具有统计学意义"
from scipy.stats import ttest_ind
ttest_ind(drug, placebo) # (非配对) t检验. P=0.025, 已经<0.05了
# ## 实验
#
# - 参与者被随机分为两组:
# - `给药组` vs. `安慰剂组`
# - 测量参与者的IQ分数
# ## 先验知识
# - 被测数据符合t分布: $ data \sim StudentsT(\mu, \sigma, \nu) $
#
# 以下为t分布的几个参数:
# - 均值符合正态分布: $ \mu \sim N(0, 100^2) $
# - 自由度(degrees of freedom)符合指数分布: $ \nu \sim Exp(30) $
# - 方差是positively-distributed: $ \sigma \sim HalfCauchy(100^2) $
# 
# ## 数据
# In[19]:
fig = plot_drug()
plt.show()
# ## 代码
# In[20]:
y_vals = np.concatenate([drug, placebo])
labels = ['drug'] * len(drug) + ['placebo'] * len(placebo)
data = pd.DataFrame([y_vals, labels]).T
data.columns = ['IQ', 'treatment']
# In[21]:
with pm.Model() as kruschke_model:
# Focus on the use of Distribution Objects.
# Linking Distribution Objects together is done by
# passing objects into other objects' parameters.
# 标准建模动作, 用pm.Xxx指定先验知识, 也就是各个参数的分布
# 注意给药组和对照组的参数要分开单独设定,
mu_drug = pm.Normal('mu_drug', mu=0, sd=100**2)
mu_placebo = pm.Normal('mu_placebo', mu=0, sd=100**2)
sigma_drug = pm.HalfCauchy('sigma_drug', beta=100)
sigma_placebo = pm.HalfCauchy('sigma_placebo', beta=100)
nu = pm.Exponential('nu', lam=1/29) + 1
# 代入参数, 为两组的分布建模
drug_like = pm.StudentT('drug', nu=nu, mu=mu_drug,
sd=sigma_drug, observed=drug)
placebo_like = pm.StudentT('placebo', nu=nu, mu=mu_placebo,
sd=sigma_placebo, observed=placebo)
# 计算组间均值的差距
diff_means = pm.Deterministic('diff_means', mu_drug - mu_placebo)
# 这俩是啥?
pooled_sd = pm.Deterministic('pooled_sd',
np.sqrt(np.power(sigma_drug, 2) +
np.power(sigma_placebo, 2) / 2))
effect_size = pm.Deterministic('effect_size',
diff_means / pooled_sd)
# ## MCMC Inference Button (TM)
# In[22]:
with kruschke_model:
kruschke_trace = pm.sample(10000, step=pm.Metropolis()) # 标准动作, 照写
# ## 结果
# In[23]:
pm.traceplot(kruschke_trace[2000:],
varnames=['mu_drug', 'mu_placebo'])
plt.show()
# In[24]:
pm.plot_posterior(kruschke_trace[2000:], color='#87ceeb',
varnames=['mu_drug', 'mu_placebo', 'diff_means'])
plt.show()
# - IQ均值的差距为: [0.5, 4.6]
# - 频率主义的 p-value: $ 0.02 $ (!!!!!!!!)
#
# 注: IQ的差异在10以上才有点意义. p-value=0.02说明组间有差异, 但没说差异有多大. 这个故事说的是虽然有差异, 但是差异太小了, 也没啥意思.
# In[25]:
def get_forestplot_line(ax, kind):
widths = {'median': 2.8, 'iqr': 2.0, 'hpd': 1.0}
assert kind in widths.keys() #f'line kind must be one of {widths.keys()}'
lines = []
for child in ax.get_children():
if isinstance(child, mpl.lines.Line2D) and np.allclose(child.get_lw(), widths[kind]):
lines.append(child)
return lines
def adjust_forestplot_for_slides(ax):
for line in get_forestplot_line(ax, kind='median'):
line.set_markersize(10)
for line in get_forestplot_line(ax, kind='iqr'):
line.set_linewidth(5)
for line in get_forestplot_line(ax, kind='hpd'):
line.set_linewidth(3)
return ax
# In[26]:
pm.forestplot(kruschke_trace[2000:],
varnames=['mu_drug', 'mu_placebo'])
ax = plt.gca()
ax = adjust_forestplot_for_slides(ax)
plt.show()
# **森林图**:在同一轴上的95%HPD(细线),IQR(粗线)和后验分布的中位数(点),使我们能够直接比较治疗组和对照组。
#
# In[27]:
def overlay_effect_size(ax):
height = ax.get_ylim()[1] * 0.5
ax.hlines(height, 0, 0.2, 'red', lw=5)
ax.hlines(height, 0.2, 0.8, 'blue', lw=5)
ax.hlines(height, 0.8, ax.get_xlim()[1], 'green', lw=5)
# In[28]:
ax = pm.plot_posterior(kruschke_trace[2000:],
varnames=['effect_size'],
color='#87ceeb')
overlay_effect_size(ax)
# - 效果大小(Cohen's d, <font style="color:red;">效果微小</font>, <font style="color:blue;">效果中等</font>, <font style="color:green;">效果很大</font>)可以从微小到很大(95%HPD [0.0,0.77])。
# - 智商提高0-4分。
# - 这种药很可能是无关紧要的。
# - 没有**生物学意义**的证据。
# # 例 2: 手机消毒问题
#
# 比较两种常用的消毒方法, 和我的fancy方法, 哪种消毒方法更好
# ## 实验设计
#
# - 将手机随机分到6组: 4 "fancy" 方法 + 2 "control" 方法.
# - 处理前后对手机表面进行拭子菌培养
# - **count** 菌落数量, 比较处理前后的菌落计数
# In[29]:
renamed_treatments = dict()
renamed_treatments['FBM_2'] = 'FM1'
renamed_treatments['bleachwipe'] = 'CTRL1'
renamed_treatments['ethanol'] = 'CTRL2'
renamed_treatments['kimwipe'] = 'FM2'
renamed_treatments['phonesoap'] = 'FM3'
renamed_treatments['quatricide'] = 'FM4'
# Reload the data one more time.
data = pd.read_csv('datasets/smartphone_sanitization_manuscript.csv', na_values=['#DIV/0!'])
del data['perc_reduction colonies']
# Exclude cellblaster data
data = data[data['treatment'] != 'CB30']
data = data[data['treatment'] != 'cellblaster']
# Rename treatments
data['treatment'] = data['treatment'].apply(lambda x: renamed_treatments[x])
# Sort the data according to the treatments.
treatment_order = ['FM1', 'FM2', 'FM3', 'FM4', 'CTRL1', 'CTRL2']
data['treatment'] = data['treatment'].astype('category')
data['treatment'].cat.set_categories(treatment_order, inplace=True)
data['treatment'] = data['treatment'].cat.codes.astype('int32')
data = data.sort_values(['treatment']).reset_index(drop=True)
data['site'] = data['site'].astype('category').cat.codes.astype('int32')
data['frac_change_colonies'] = ((data['colonies_post'] - data['colonies_pre'])
/ data['colonies_pre'])
data['frac_change_colonies'] = pm.floatX(data['frac_change_colonies'])
del data['screen protector']
# Change dtypes to int32 for GPU usage.
def change_dtype(data, dtype='int32'):
return data.astype(dtype)
cols_to_change_ints = ['sample_id', 'colonies_pre', 'colonies_post',
'morphologies_pre', 'morphologies_post', 'phone ID']
cols_to_change_floats = ['year', 'month', 'day', 'perc_reduction morph',
'phone ID', 'no case',]
for col in cols_to_change_ints:
data[col] = change_dtype(data[col], dtype='int32')
for col in cols_to_change_floats:
data[col] = change_dtype(data[col], dtype='float32')
data.dtypes
# # filter the data such that we have only PhoneSoap (PS-300) and Ethanol (ET)
# data_filtered = data[(data['treatment'] == 'PS-300') | (data['treatment'] == 'QA')]
# data_filtered = data_filtered[data_filtered['site'] == 'phone']
# data_filtered.sample(10)
# ## 数据
# In[ ]:
def plot_colonies_data():
fig = plt.figure(figsize=(10,5))
ax1 = fig.add_subplot(2,1,1)
sns.swarmplot(x='treatment', y='colonies_pre', data=data, ax=ax1)
ax1.set_title('pre-treatment')
ax1.set_xlabel('')
ax1.set_ylabel('colonies')
ax2 = fig.add_subplot(2,1,2)
sns.swarmplot(x='treatment', y='colonies_post', data=data, ax=ax2)
ax2.set_title('post-treatment')
ax2.set_ylabel('colonies')
ax2.set_ylim(ax1.get_ylim())
plt.tight_layout()
return fig
# In[ ]:
fig = plot_colonies_data()
plt.show()
# ## 先验知识
#
# 菌落计数符合**泊松Poisson分布**. 因此...
#
# - 菌落计数符合泊松分布: $ data_{i}^{j} \sim Poisson(\mu_{i}^{j}), j \in [pre, post], i \in [1, 2, 3...] $
# - 泊松分布的参数是离散均匀分布: $ \mu_{i}^{j} \sim DiscreteUniform(0, 10^{4}), j \in [pre, post], i \in [1, 2, 3...] $
# - 灭菌效力通过百分比变化测量,定义如下: $ \frac{mu_{pre} - mu_{post}}{mu_{pre}} $
# 
# ## 代码
# In[ ]:
with pm.Model() as poisson_estimation:
mu_pre = pm.DiscreteUniform('pre_mus', lower=0, upper=10000,
shape=len(treatment_order))
pre_mus = mu_pre[data['treatment'].values] # fancy indexing!!
pre_counts = pm.Poisson('pre_counts', mu=pre_mus,
observed=pm.floatX(data['colonies_pre']))
mu_post = pm.DiscreteUniform('post_mus', lower=0, upper=10000,
shape=len(treatment_order))
post_mus = mu_post[data['treatment'].values] # fancy indexing!!
post_counts = pm.Poisson('post_counts', mu=post_mus,
observed=pm.floatX(data['colonies_post']))
perc_change = pm.Deterministic('perc_change',
100 * (mu_pre - mu_post) / mu_pre)
# ## MCMC Inference Button (TM)
# In[ ]:
with poisson_estimation:
poisson_trace = pm.sample(200000)
# In[ ]:
pm.traceplot(poisson_trace[50000:], varnames=['pre_mus', 'post_mus'])
plt.show()
# ## 结果
# In[ ]:
pm.forestplot(poisson_trace[50000:], varnames=['perc_change'],
ylabels=treatment_order) #, xrange=[0, 110])
plt.xlabel('Percentage Reduction')
ax = plt.gca()
ax = adjust_forestplot_for_slides(ax)
# # 第三类问题: 复杂的东西
# # 例子: 贝叶斯神经网络
#
# a.k.a. 贝叶斯深度学习
# 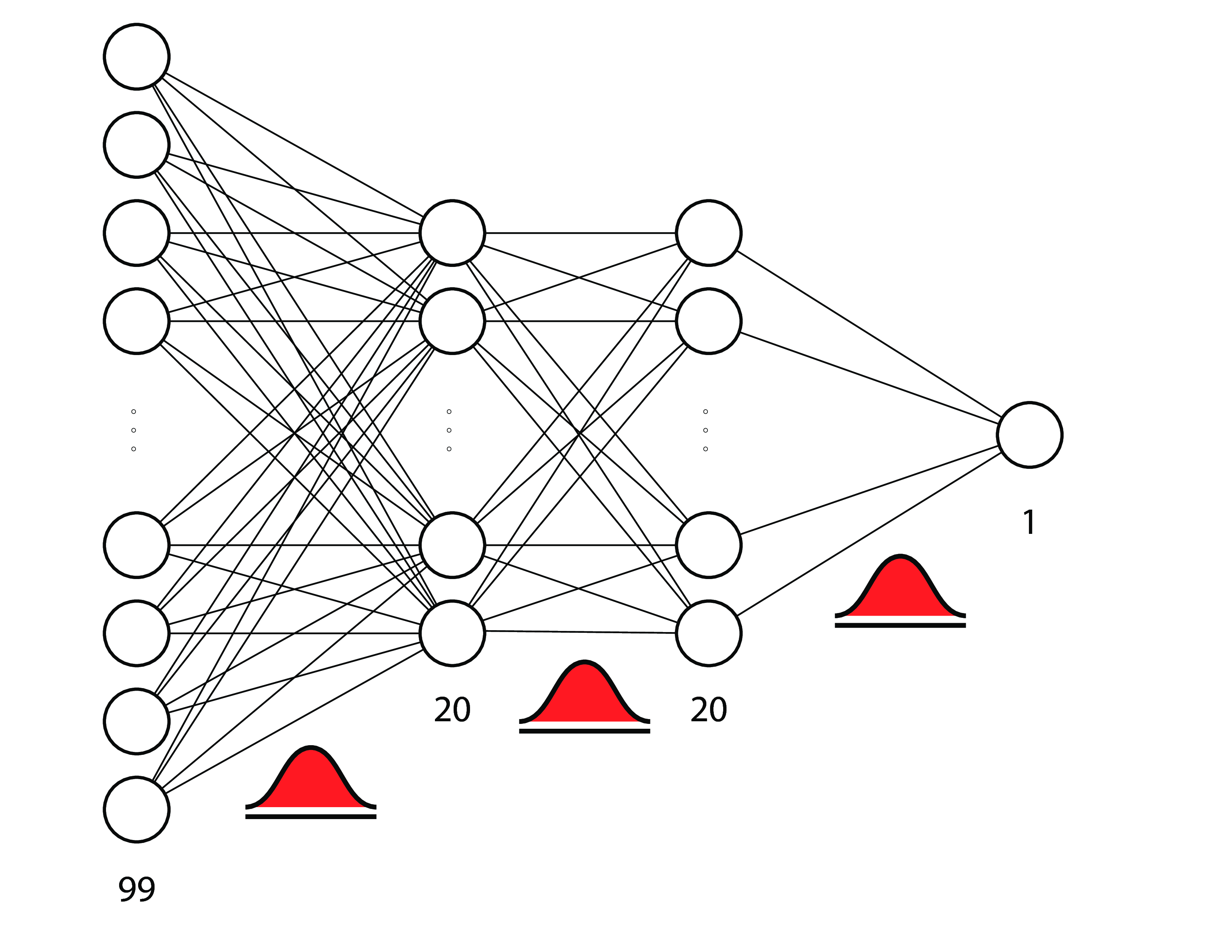
# [Forest Cover Notebook](https://github.com/ericmjl/bayesian-analysis-recipes/blob/master/multiclass-classification-neural-network.ipynb)
#
# 注: 这好像跳到另一个课件去了. 有时间我也挪过来翻译
# # 概念特征
#
# - 参数估计:
# - **抛硬币:** 先验与后验
# - ** IC <sub> 50 </ sub>:**连接函数与确定性计算
# - 对照与治疗:
# - **药物智商:**一个治疗组 与 一个对照组
# - **电话消毒:**多个治疗组 与 多个对照组。
# - 贝叶斯神经网络:
# - **森林覆盖:**先验参数和大致推断。
# # 模式
#
# 1. 使用统计分布参数化您的问题
# 1. 修正你的模型结构
# 1. 在PyMC3中编写模型,点击**Inference 按钮<sup>TM</sup>**
# 1. 根据后验分布进行解释
# 1. (可选)如果有新信息,修改模型结构。
# # 贝叶斯估计
#
# - 为数据的生成写一个**描述性的**模型。
# - 原始的贝叶斯:在**看到你的数据之前** 做这个。
# - 经验贝叶斯:在**看到你的数据之后**做这个。
# - 估计感兴趣的模型参数的**后验分布**。
# - **确定性计算** 派生参数的后验分布。
# # 参考资源
#
# - John K. Kruschke's [books][kruschke_books], [paper][kruschke_paper], and [video][kruschke_video].
# - Statistical Re-thinking [book][mcelreath]
# - Jake Vanderplas' [blog post][jakevdp_blog] on the differences between Frequentism and Bayesianism.
# - PyMC3 [examples & documentation][pymc3]
# - Andrew Gelman's [blog][gelman]
# - Recommendations for prior distributions [wiki][priors]
# - Cam Davidson-Pilon's [Bayesian Methods for Hackers][bayes_hacks]
# - My [repository][bayes_recipes] of Bayesian data analysis recipes.
#
# [kruschke_books]: https://sites.google.com/site/doingbayesiandataanalysis/
# [kruschke_paper]: http://www.indiana.edu/~kruschke/BEST/
# [kruschke_video]: https://www.youtube.com/watch?v=fhw1j1Ru2i0&feature=youtu.be
# [jakevdp_blog]: http://jakevdp.github.io/blog/2014/03/11/frequentism-and-bayesianism-a-practical-intro/
# [pymc3]: https://pymc-devs.github.io/pymc3/examples.html
# [mcelreath]: http://xcelab.net/rm/statistical-rethinking/
# [gelman]: http://andrewgelman.com/
# [priors]: https://github.com/stan-dev/stan/wiki/Prior-Choice-Recommendations
# [bayes_hacks]: https://github.com/CamDavidsonPilon/Probabilistic-Programming-and-Bayesian-Methods-for-Hackers
# [bayes_recipes]: https://github.com/ericmjl/bayesian-analysis-recipes
# # GO BAYES!
#
# - Full notebook with bonus resources: https://github.com/ericmjl/bayesian-stats-talk
# - Twitter: [@ericmjl](https://twitter.com/ericmjl)
# - Website: [ericmjl.com](http://www.ericmjl.com)