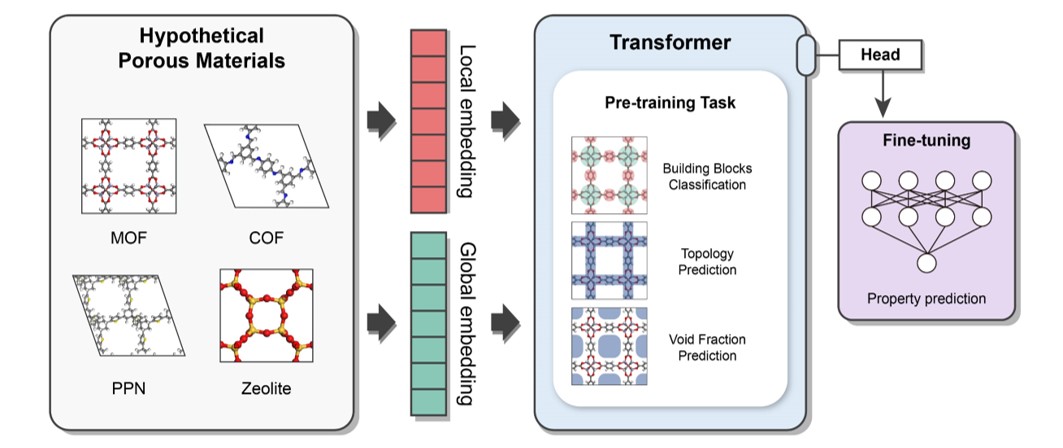
This package provides a universal transfer learning model, PMTransformer (Porous Materials Transformer), which obtains the state-of-the-art performance in predicting various properties of porous materials. The PMTRansformer was pre-trainied with 1.9 million hypothetical porous materials including Metal-Organic Frameworks (MOFs), Covalent-Organic Frameworks (COFs), Porous Polymer Networks (PPNs), and zeolites. By fine-tuning the pre-trained PMTransformer, you can easily obtain machine learning models to accurately predict various properties of porous materials .
NOTE: From version 2.0.0, the default pre-training model has been changed from MOFTransformer to PMTransformer, which was pre-trained with a larger dataset, containing other porous materials as well as MOFs. The PMTransformer outperforms the MOFTransformer in predicting various properties of porous materials.
Version: 2.2.0
Now, MOFTransformer support multi-task learning (see multi-task learning)
NOTE: This package is primarily tested on Linux. We strongly recommend using Linux for the installation.
python>=3.8
Given that MOFTransformer is based on pytorch, please install pytorch (>= 1.12.0) according to your environments.
$ pip install moftransformer
- you can download the pretrained models (
PMTransformer.ckptandMOFTransformer.ckpt) via figshare
or you can download with a command line:
$ moftransformer download pretrain_model
- we've provide the pre-embeddings (i.e., atom-based graph embeddings and energy-grid embeddings), inputs of
PMTransformer, for CoREMOF, QMOF database.
$ moftransformer download coremof
$ moftransformer download qmof
- Install
GRIDAYto calculate energy-grids from cif files
$ moftransformer install-griday
- Run prepare-data .
from moftransformer.examples import example_path
from moftransformer.utils import prepare_data
# Get example path
root_cifs = example_path['root_cif']
root_dataset = example_path['root_dataset']
downstream = example_path['downstream']
train_fraction = 0.8 # default value
test_fraction = 0.1 # default value
# Run prepare data
prepare_data(root_cifs, root_dataset, downstream=downstream,
train_fraction=train_fraction, test_fraction=test_fraction)- Fine-tune the pretrained MOFTransformer.
import moftransformer
from moftransformer.examples import example_path
# data root and downstream from example
root_dataset = example_path['root_dataset']
downstream = example_path['downstream']
log_dir = './logs/'
# load_path = "pmtransformer" (default)
# kwargs (optional)
max_epochs = 10
batch_size = 8
mean = 0
std = 1
moftransformer.run(root_dataset, downstream, log_dir=log_dir,
max_epochs=max_epochs, batch_size=batch_size,
mean=mean, std=std)- Test fine-tuned model
from pathlib import Path
import moftransformer
from moftransformer.examples import example_path
root_dataset = example_path['root_dataset']
downstream = example_path['downstream']
# Get ckpt file
seed = 0 # default seeds
version = 0 # version for model. It increases with the number of trains
# For version > 2.1.1, best.ckpt exists
checkpoint = 'best' # Epochs where the model is stored.
save_dir = 'result/'
# optional keyword
mean = 0
std = 1
load_path = Path(log_dir) / f'pretrained_mof_seed{seed}_from_pmtransformer/version_{version}/checkpoints/{checkpoint}.ckpt'
if not load_path.exists():
raise ValueError(f'load_path does not exists. check path for .ckpt file : {load_path}')
moftransformer.test(root_dataset, load_path, downstream=downstream,
save_dir=save_dir, mean=mean, std=std)- predict from fine-tuned model
from pathlib import Path
import moftransformer
from moftransformer.examples import example_path
root_dataset = example_path['root_dataset']
downstream = example_path['downstream']
# Get ckpt file
log_dir = './logs/' # same directory make from training
seed = 0 # default seeds
version = 0 # version for model. It increases with the number of trains
checkpoint = 'best' # Epochs where the model is stored.
mean = 0
std = 1
load_path = Path(log_dir) / f'pretrained_mof_seed{seed}_from_pmtransformer/version_{version}/checkpoints/{checkpoint}.ckpt'
if not load_path.exists():
raise ValueError(f'load_path does not exists. check path for .ckpt file : {load_path}')
moftransformer.predict(
root_dataset, load_path=load_path, downstream=downstream, split='all', mean=mean, std=std
)- Visualize analysis of feature importance for the fine-tuned model. (You should download or train
fine-tunedmodel before visualization)
from moftransformer.visualize import PatchVisualizer
from moftransformer.examples import visualize_example_path
model_path = "examples/finetuned_bandgap.ckpt" # or 'examples/finetuned_h2_uptake.ckpt'
data_path = visualize_example_path
cifname = 'MIBQAR01_FSR'
vis = PatchVisualizer.from_cifname(cifname, model_path, data_path)
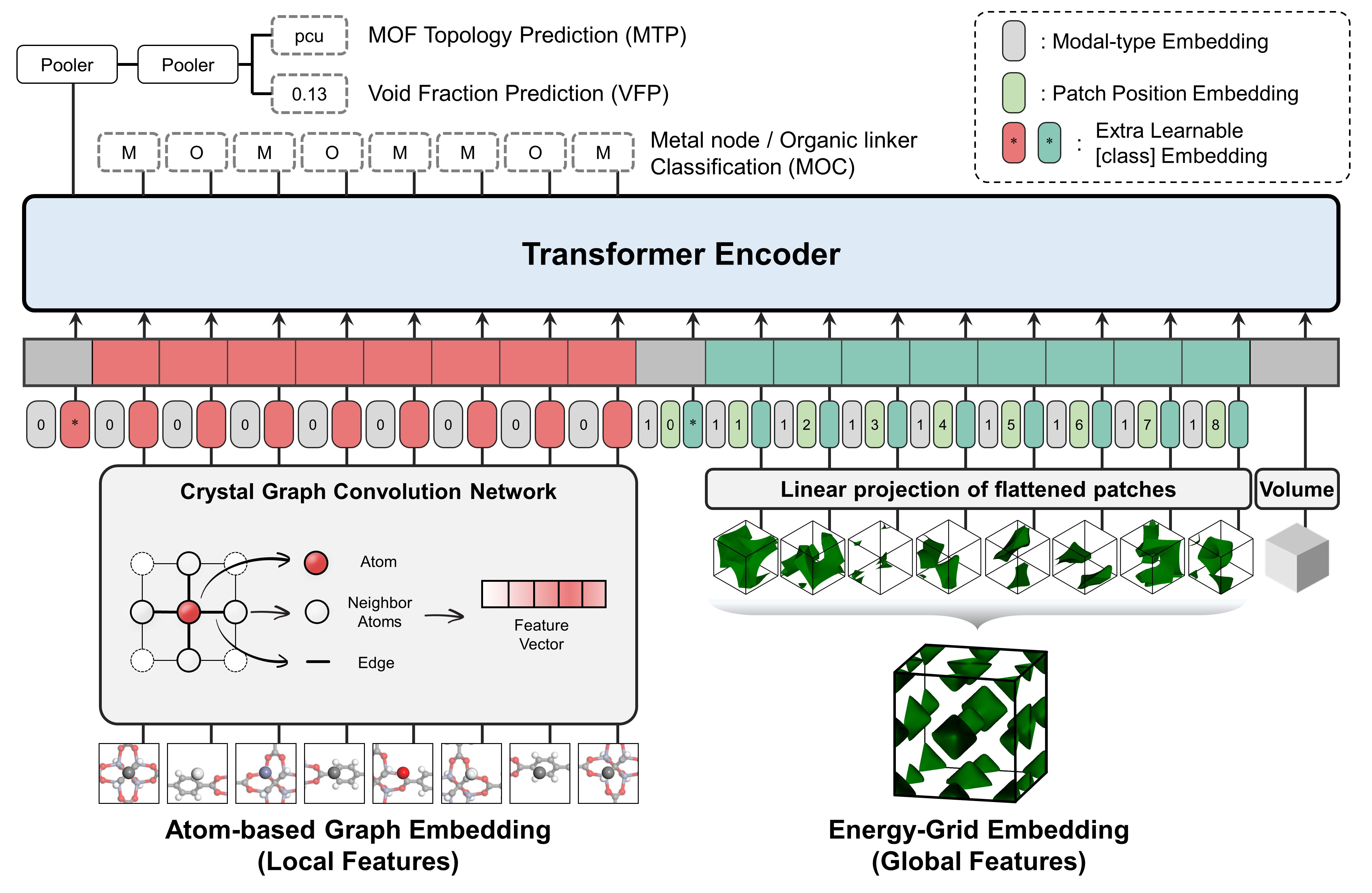
vis.draw_graph()It is a multi-modal pre-training Transformer encoder which is designed to capture both local and global features of porous materials.
The pre-traning tasks are as follows: (1) Topology Prediction (2) Void Fraction Prediction (3) Building Block Classification
It takes two different representations as input
- Atom-based Graph Embedding : CGCNN w/o pooling layer -> local features
- Energy-grid Embedding : 1D flatten patches of 3D energy grid -> global features
you can easily visualize feature importance analysis of atom-based graph embeddings and energy-grid embeddings.
%matplotlib widget
from visualize import PatchVisualizer
model_path = "examples/finetuned_bandgap.ckpt" # or 'examples/finetuned_h2_uptake.ckpt'
data_path = 'examples/visualize/dataset/'
cifname = 'MIBQAR01_FSR'
vis = PatchVisualizer.from_cifname(cifname, model_path, data_path)
vis.draw_graph()vis = PatchVisualizer.from_cifname(cifname, model_path, data_path)
vis.draw_grid()Comparison of mean absolute error (MAE) values for various baseline models, scratch, MOFTransformer, and PMTransformer on different properties of MOFs, COFs, PPNs, and zeolites. The bold values indicate the lowest MAE value for each property. The details of information can be found in PMTransformer paper
| Material | Property | Number of Dataset | Energy histogram | Descriptor-based ML | CGCNN | Scratch | MOFTransformer | PMTransformer |
|---|---|---|---|---|---|---|---|---|
| MOF | H2 Uptake (100 bar) | 20,000 | 9.183 | 9.456 | 32.864 | 7.018 | 6.377 | 5.963 |
| MOF | H2 diffusivity (dilute) | 20,000 | 0.644 | 0.398 | 0.6600 | 0.391 | 0.367 | 0.366 |
| MOF | Band-gap | 20.373 | 0.913 | 0.590 | 0.290 | 0.271 | 0.224 | 0.216 |
| MOF | N2 uptake (1 bar) | 5,286 | 0.178 | 0.115 | 0.108 | 0.102 | 0.071 | 0.069 |
| MOF | O2 uptake (1 bar) | 5,286 | 0.162 | 0.076 | 0.083 | 0.071 | 0.051 | 0.053 |
| MOF | N2 diffusivity (1 bar) | 5,286 | 7.82e-5 | 5.22e-5 | 7.19e-5 | 5.82e-05 | 4.52e-05 | 4.53e-05 |
| MOF | O2 diffusivity (1 bar) | 5,286 | 7.14e-5 | 4.59e-5 | 6.56e-5 | 5.00e-05 | 4.04e-05 | 3.99e-05 |
| MOF | CO2 Henry coefficient | 8,183 | 0.737 | 0.468 | 0.426 | 0.362 | 0.295 | 0.288 |
| MOF | Thermal stability | 3,098 | 68.74 | 49.27 | 52.38 | 52.557 | 45.875 | 45.766 |
| COF | CH4 uptake (65bar) | 39,304 | 5.588 | 4.630 | 15.31 | 2.883 | 2.268 | 2.126 |
| COF | CH4 uptake (5.8bar) | 39,304 | 3.444 | 1.853 | 5.620 | 1.255 | 0.999 | 1.009 |
| COF | CO2 heat of adsorption | 39,304 | 2.101 | 1.341 | 1.846 | 1.058 | 0.874 | 0.842 |
| COF | CO2 log KH | 39,304 | 0.242 | 0.169 | 0.238 | 0.134 | 0.108 | 0.103 |
| PPN | CH4 uptake (65bar) | 17,870 | 6.260 | 4.233 | 9.731 | 3.748 | 3.187 | 2.995 |
| PPN | CH4 uptake (1bar) | 17,870 | 1.356 | 0.563 | 1.525 | 0.602 | 0.493 | 0.461 |
| Zeolite | CH4 KH (unitless) | 99,204 | 8.032 | 6.268 | 6.334 | 4.286 | 4.103 | 3.998 |
| Zeolite | CH4 Heat of adsorption | 99,204 | 1.612 | 1.033 | 1.603 | 0.670 | 0.647 | 0.639 |
if you want to cite PMTransformer or MOFTransformer, please refer to the following paper:
-
A multi-modal pre-training transformer for universal transfer learning in metal–organic frameworks, Nature Machine Intelligence, 5, 2023. link
-
Enhancing Structure–Property Relationships in Porous Materials through Transfer Learning and Cross-Material Few-Shot Learning, ACS Appl. Mater. Interfaces 2023, 15, 48, 56375–56385. link
Contributions are welcome! If you have any suggestions or find any issues, please open an issue or a pull request.
This project is licensed under the MIT License. See the LICENSE file for more information.