-
Notifications
You must be signed in to change notification settings - Fork 0
/
Copy pathhandout.Rmd
1022 lines (814 loc) · 37.6 KB
/
handout.Rmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
398
399
400
401
402
403
404
405
406
407
408
409
410
411
412
413
414
415
416
417
418
419
420
421
422
423
424
425
426
427
428
429
430
431
432
433
434
435
436
437
438
439
440
441
442
443
444
445
446
447
448
449
450
451
452
453
454
455
456
457
458
459
460
461
462
463
464
465
466
467
468
469
470
471
472
473
474
475
476
477
478
479
480
481
482
483
484
485
486
487
488
489
490
491
492
493
494
495
496
497
498
499
500
501
502
503
504
505
506
507
508
509
510
511
512
513
514
515
516
517
518
519
520
521
522
523
524
525
526
527
528
529
530
531
532
533
534
535
536
537
538
539
540
541
542
543
544
545
546
547
548
549
550
551
552
553
554
555
556
557
558
559
560
561
562
563
564
565
566
567
568
569
570
571
572
573
574
575
576
577
578
579
580
581
582
583
584
585
586
587
588
589
590
591
592
593
594
595
596
597
598
599
600
601
602
603
604
605
606
607
608
609
610
611
612
613
614
615
616
617
618
619
620
621
622
623
624
625
626
627
628
629
630
631
632
633
634
635
636
637
638
639
640
641
642
643
644
645
646
647
648
649
650
651
652
653
654
655
656
657
658
659
660
661
662
663
664
665
666
667
668
669
670
671
672
673
674
675
676
677
678
679
680
681
682
683
684
685
686
687
688
689
690
691
692
693
694
695
696
697
698
699
700
701
702
703
704
705
706
707
708
709
710
711
712
713
714
715
716
717
718
719
720
721
722
723
724
725
726
727
728
729
730
731
732
733
734
735
736
737
738
739
740
741
742
743
744
745
746
747
748
749
750
751
752
753
754
755
756
757
758
759
760
761
762
763
764
765
766
767
768
769
770
771
772
773
774
775
776
777
778
779
780
781
782
783
784
785
786
787
788
789
790
791
792
793
794
795
796
797
798
799
800
801
802
803
804
805
806
807
808
809
810
811
812
813
814
815
816
817
818
819
820
821
822
823
824
825
826
827
828
829
830
831
832
833
834
835
836
837
838
839
840
841
842
843
844
845
846
847
848
849
850
851
852
853
854
855
856
857
858
859
860
861
862
863
864
865
866
867
868
869
870
871
872
873
874
875
876
877
878
879
880
881
882
883
884
885
886
887
888
889
890
891
892
893
894
895
896
897
898
899
900
901
902
903
904
905
906
907
908
909
910
911
912
913
914
915
916
917
918
919
920
921
922
923
924
925
926
927
928
929
930
931
932
933
934
935
936
937
938
939
940
941
942
943
944
945
946
947
948
949
950
951
952
953
954
955
956
957
958
959
960
961
962
963
964
965
966
967
968
969
970
971
972
973
974
975
976
977
978
979
980
981
982
983
984
985
986
987
988
989
990
991
992
993
994
995
996
997
998
999
1000
---
title: "Introduction to R"
author: "Jacob Simmering"
date: "Nov 1st, 2017"
output:
html_document: default
---
```{r setup, include=FALSE}
knitr::opts_chunk$set(echo = TRUE)
library(tidyverse)
```
# Setup
You will need to have a version of R installed. It can be downloaded for your
system at [CRAN](https://cran.r-project.org/). Also you will want to install
the `tidyverse` inside R. You can do this by running
`install.packages("tidyverse")` in an R session.
Finally, while not required, the [RStudio IDE](https://www.rstudio.com/) will
make your R sessions much
easier to manage.
# Overview
## The R Langague
I am not going to spend a lot of time R as a language. I'll cover some syntax
as we reach it. I'll cover just a few brief terms and conventions here.
*Packages* are contributed extensions to the R language to enable specific
tasks or analyses. If you are familiar with Python, these are like libraries or
gems in Ruby. Most of the time, there is a contributed version on CRAN that you
can easily install with `install.packages("<package name>")` in your R session.
It will download and install the requested package as well as any dependencies
required. Once a package is installed, you can load it into your session with
`library(<package name>)`. Quotes are required to install but optional when
loading the library.
The `<-` (read as "gets") is the primary assignment operator in R. If you want
to set the value of `x` to `1`, you would run `x <- 1`. This is directional,
so `1 -> x` is also valid (useful if you just typed a long line and forgot
to assign it, but probably not good practice!). You can use `=` for assignment
(and indeed need to when you are passing function arguments) but for whatever
reason that is not standard practice.
The operator `%>%` is added when you load the `tidyverse` package and is known
as the "pipe" operator. You can understand read this as "then". This is used
to string together a series of operations into one group while splitting across
lines for readability and avoiding nesting of parenthesis. We'll discuss this
later in context.
There are three atomic data types in R.
* "numeric" - this stores floating point and integer values. Typically, it will
store the value as a double precision floating point but integer can be
forced if desired.
* "factor" - this looks like a string but it isn't! Factors are categorical
variables with discrete levels (e.g., male vs female, dog vs cat vs mouse).
In R, they are actually stored internally as a series of integers and R converts
these integers to labels (e.g., 1 = male, 2 = female) when it shows output.
This can be confusing because they look like strings but aren't - it makes sense
in R's standard statistical use-case but will almost certainly make everyone
groan at least once.
* "character" - this is actually the string-type in R. Most of the tools I'll
discuss today don't convert characters to factors but historically R has
coerced characters into factors when reading data into a session.
There are a set of higher level data types in R:
* "vector" - a vector is 1-D collection of values, all of the same data type.
A "vector" in R and a "vector" in math mean approximately the same thing.
For instance `c(1, 2, 3)` is a vector as is `c("A", "B", "C")` (`c()` is the
function used to construct a vector, `c` standards for "combine" or
"concatenation"). R will automatically convert a vector with mixed types into
the most strict atomic data type that can hold all of the values in the vector.
For instance, `c(1, 2, "A")` will become a string of character values.
* "matrix" - a matrix is a 2-D collection of values. This is the same as a
"matrix" in math. Like a vector, it can only hold one atomic data type.
* "array" - an array is a generalization of a matrix to any number of
dimensions. This can be useful to hold data where you have clear "slices"
(e.g., intensity by x and y at several difference values of t). Again, this can
only hold one atomic data type.
* "data.frame" - a data.frame is the primary data storage object in base R. It
is a collection of vectors, each displayed as a column, and is analogous to
a table or spreadsheet. The columns can be of different types.
* "tibble" - a tibble (a corruption of the word "table" which was abbreviated as
"tbl" in the output) is an extension of the data.frame provided by the `tibble`
package in the `tidyverse`. The main advantages of a tibble are a better print
method and it makes it a little harder to get yourself in trouble. For more
on tibbles see [R for Data Science Chapter 10](http://r4ds.had.co.nz/tibbles.html).
* "list" - a list is a collection of any R data types with each element being
potentially a different type. These come in handy when doing functional
programming tasks.
I am going to be mostly using tibble type objects here. We will touch briefly
on lists towards the end if time permits. I'll mention vectors in a few places
as they will be used next week.
## The Packages We'll Use Today
I am going to cover using R, mostly focusing on data importation, manipulation
and visualization. I'll also cover the different data types in R, extensions of
these data types and, time permitting, some functional programming in R. Next
week Aaron Miller will cover machine learning in R.
Most of the examples in this book will use a package called `tidyverse`. The
tidyverse is a collection of tools to help with the data importation and
manipulation challenges in R. Specifically, we will discuss the following
packages this week
* `dplyr`
* `ggplot2`
* `tidyr`
* `readr`
* `stringr`
* `purrr`
* `broom`
## The Data We'll Use Today
We will be using the data sets in the `nycflights13` package. However, instead
of using the package we are going to read in the data directly. We'll discuss
reading in the data next, but first a brief overview of the data. The data is
a summary of the on-time data for all flights that left NYC in 2013 and
includes "metadata" such as airline, airport the plane was flying to, weather
and the plane. The data was collected and compiled into the R package by
Hadley Wickham in 2017.
There are five tables. They are
* airlines
* airports
* flights
* planes
* weather
A sample schema showing how they are joined (taken from R4DS)
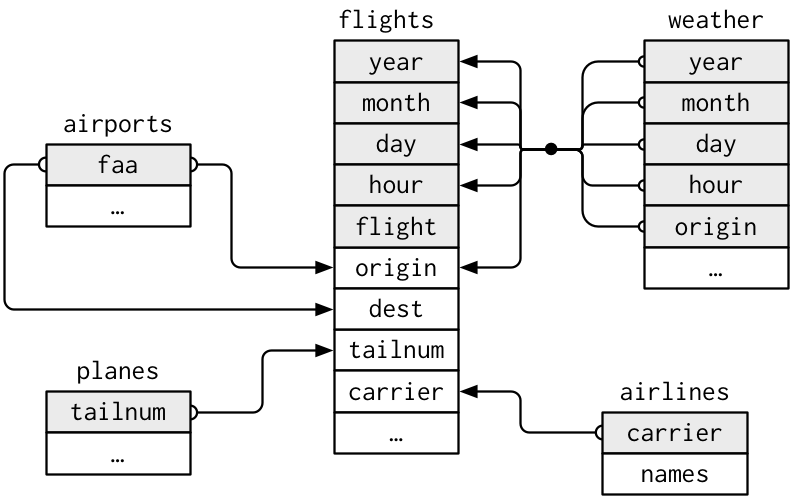
# Data Science Workflow
The tidyverse suggests a workflow of data exploration and analysis depicted
in the figure below (taken from R for Data Science)
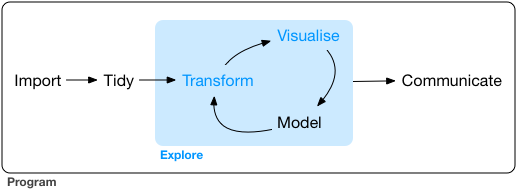
The ideal is that you import your (potentially unstructured/messy) data,
tidy it (convert it into a cleaned, rectangular format, more on tidy data later)
and then enter the analysis/exploration cycle. Here you start by transforming
your data, visualizing relationships, modeling those relationships and then
potentially returning to the transformation and repeating the process.
Once you have finalized the model/completed the analysis, you communicate
your results.
The tidyverse package has a number of child packages that cover these tasks
more-or-less in a 1:1 fashion:
* `readr` covers many forms of data import into R
* `tidyr` assists with tidying messy data and getting it into the desired
format
* `dplyr` covers many, if not most, transformations of the data you might want
to do
* `ggplot2` is a powerful and visually appealing tool for producing
visualizations of your data and any relationships within
* `modelr`, `purrr` and the base R functions `lm()` and `glm()` form the bulk of
the modeling stage
* `knitr` provides an easy way to embedded R code and results inside Markdown
files for easy drafting of reports and communication
I will briefly cover the use of `readr` and `tidyr` today. I will cover
`dplyr` in-depth and provide an introduction to `ggplot2`. There won't be time
to cover the regression and model methods in much detail without getting bogged
down in the stats
# Data Import
For more information see
[Chapter 11 of R for Data Science](http://r4ds.had.co.nz/data-import.html)
Start a new R script in RStudio and add the following to the top of the script
```{r eval=FALSE}
library(tidyverse)
```
And run the line (click on the "Run" icon or put your cursor on the line and
press crtl-enter (Windows, Linux) or cmd-enter (OS X)). This should send your
code to the console to execute.
If you see the message
`Error in library(tidyverse) : there is no package called 'tidyverse'` it means
that the `tidyverse` package is not installed. In the console, run
`install.packages("tidyverse")` to install it and then re-run the line.
If you see the following:
```
Loading tidyverse: ggplot2
Loading tidyverse: tibble
Loading tidyverse: tidyr
Loading tidyverse: readr
Loading tidyverse: purrr
Loading tidyverse: dplyr
Conflicts with tidy packages ---------------------------------------------------
filter(): dplyr, stats
lag(): dplyr, stats
```
It means that you have successfully loaded the `tidyverse` and related packages
into your R session. The `Conflicts with tidy packages` section lists places
where there are two functions with the same name being loaded into the global
namespace. By default, the last function loaded will be the default. For
instance, if I run the function `lag()` it will use the `lag()` as defined by
the `dplyr` package since I loaded that after the `stats` package was loaded.
If I want to access the `stats` version of `lag()`, I can explicitly identify
the namespace using the format `namespace::function()` or `stats::lag()` in
this case.
We are going to use the `readr` package to load our data. There are specialized
functions in the `readr` package to read tab-separated files, fixed-with files,
delimited files and comma-separated files. We are going to focus on csv's, but
you can read in pretty much any plain-text file with `readr`.
The main function we want to use is `read_csv()`. If you enter `?read_csv` in
the console, you should be able to see the Help package for the `read_csv()`
function. You'll see the description that: "read_csv() and read_tsv() are
special cases of the general read_delim(). They're useful for reading the most
common types of flat file data, comma separated values and tab separated values,
respectively. read_csv2() uses ; for separators, instead of ,. This is common in
European countries which use , as the decimal separator." Looking under the
Usage entry, we see
```
read_csv(file, col_names = TRUE, col_types = NULL,
locale = default_locale(), na = c("", "NA"), quoted_na = TRUE,
quote = "\"", comment = "", trim_ws = TRUE, skip = 0, n_max = Inf,
guess_max = min(1000, n_max), progress = show_progress())
```
That is a lot of arguments to manage and the arguments section goes through
them in detail. We won't need to change many of these from defaults. In general,
you only need to worry about setting `file` and, occasionally, `col_types`.
I've provided all of the data we are going to use in the `data` directory of
this repo. Let's one of the files and print it:
```{r data_load_1}
airlines <- read_csv("data/airlines.csv")
airlines
```
The section that reads
```
## Parsed with column specification:
## cols(
## carrier = col_character(),
## name = col_character()
## )
```
tells us about the data, specifically, the variables the parser found and their
types. In this case, there are two variables, carrier and name, both of which
are characters.
When we print out the object, we see that it is a tibble with 2 columns and
16 rows. `carrier` appears to be a short 2-letter abbreviation for an airline
and `name` is the full name of the airline company.
We can do this with the rest of the files as well:
```{r data_load_2}
airports <- read_csv("data/airports.csv")
planes <- read_csv("data/planes.csv")
flights <- read_csv("data/flights.csv")
weather <- read_csv("data/weather.csv")
```
We again see a list of all the variables in the data set and their types. You'll
notice that even more complicated types, such as datetime, were recognized and
parsed.
The other data import functions in `readr` will produce similar results.
# Data Manipulation
## A Note on Tidy Data
I'm going to take a brief minute here to discuss "tidy" data. The idea behind
tidy data is similar to normalization in databases and indeed tidy data can be
thought of as a re-casting of 3rd Normal Form in a statistical language.
Specifically,
1. Each variable must have its own column;
2. Each observation must have its own row;
3. Each value must have its own cell.
If the data is stored as such, it makes manipulating it and producing summaries,
aggregations and other data transformations easier. It also means the data is
stored consistently and do you don't have to re-learn the data structure every
project.
The tools in the tidyverse are designed around the idea of tidy data and
integrate neatly with the concept. If you have tidy data, use of these tools
will feel natural and intuitive.
All of the data sets that we have are tidy, but if they aren't (or you just
need to do it) the functions `gather()` and `spread()` from the `tidyr`
package (part of the tidyverse) are useful. `gather()` takes "wide" data and
makes it long. For instance,
```{r spread_and_gather}
stocks <- tibble(
time = as.Date('2009-01-01') + 0:9,
X = rnorm(10, 0, 1), # just generates some random numbers
Y = rnorm(10, 0, 2), # just generates some random numbers
Z = rnorm(10, 0, 4) # just generates some random numbers
)
stocks
```
is the closing price of 3 stocks "X", "Y" and "Z" at the end of each day
from 2009-1-1 to 2009-1-10. If we wanted them to be long such that there
was 3 rows for each time, a column that indicates the stock and a column that
indicates the closing price, we would use `gather()`. `gather()` typically takes
4 arguments: `data` is the name of the tibble to perform the operation on,
`key` is the name to use for the column that will become the key, `value`
is the name to use for the column that will hold the value then a comma
separated list of variables to include (or prefix with a `-` sign to exclude).
For the `stocks` example
```{r gather_1}
stocks_wide <- gather(stocks, stock, closing_price, -time)
stocks_wide
```
and we can convert back to wide using `spread()`. This reverses the action of
`gather()`. The first argument should be the name of the tibble to act on
(in this case `stocks_wide`), the second should be the name of the column that
contains the values to use as the new column names (`stocks`) and the third
should contain the values used to fill the new cells (`closing_price`).
```{r}
spread(stocks_wide, stock, closing_price)
```
These are handy little tools to keep in your tool kit. Moving from wide-to-long
happens more than you'd expect.
For more on tidy data, its importance and how it works influences the
`tidyverse` see [R for Data Science Chapter 12](http://r4ds.had.co.nz/tidy-data.html).
## The `dplyr` Verbs
`dplyr` is a workhorse package to manipulating and summarizing data. The most
exciting thing about `dplyr` is that the syntax and code is the same for
working on an in-memory dataset (like we do here) or a SQL database. `dbplyr`
is a package that extends `dplyr` and converts your R code into SQL queries
and passes them off to the RDBMS. This is really cool when you are working with
large data sets and makes the transition between in-memory data manipulation and
on-disk data manipulation seamless. It is also very easy to use, which doesn't
hurt at all!
In general, `dplyr` is simply a collection of six verbs (or five verbs and one
adverb) which will do about 99% of all data manipulation tasks you might want to
do. The verbs are
* `select()`
* `filter()`
* `mutate()`
* `summarize()`
* `arrange()`
* `group_by()` (this might actually be an adverb)
We'll go through what each function does and how to use it using the
`nycflights13` data sets as an example.
### `select()`
Select does what it says - it selects things. You give it the name of a data
object and then a comma separated list of variables and it returns the data
object with only those variables.
Let's start with the `airports` table
```{r vars_in_flights}
airports
```
We might only want the `air_time` (flight time) and `distance` in our dataset.
The rest of the variables are just taking up space. We can use `select()` to
do that for us
```{r select_1}
select(airports, faa, lat, lon)
```
We can also exclude specific variables by putting a `-` in front of the variable
name
```{r select_2}
select(airports, -faa)
```
We can also use a set of helper functions to select the desired variables:
* `starts_with(match)` - returns any variables that start with the value passed
as `match`
* `ends_with(match)` - returns any variables that ends with the value passed as
`match`
* `contains(match)` - returns any variables that contain the value `match`
#### Exercises
1. Arrange the data by the distance covered by the flight
### `filter()`
`filter()` filters things. It takes as a first argument the name of the data
object (a tibble or data.frame) to filter and then a series of comma separated
expressions that evaluate to logical TRUE/FALSE statements about a given row.
The `flights` table is a good place to try this:
```{r filter_1}
flights
```
Suppose we wanted to find it any flight from NYC went to Des Moines airport
in 2013. The expression would test if `dest == "DSM"`. So
```{r filter_2}
filter(flights, dest == "DSM")
```
Suppose we wanted multiple filtering rules, we can string them together with
commas and `filter()` understands that as "AND". So `filter(data, x, y)` means
return rows in data where both x AND y are true.
```{r filter_3}
filter(flights, year == 2013, month == 1, day == 1)
```
You can join using "OR" as well. In R, the logical operator OR is expressed as
`|`. For instance, if I want to find flights that left either more than 5
minutes before or 5 minutes after their scheduled time, I would run
```{r filter_4}
filter(flights, (dep_time - sched_dep_time) >= 5 | (dep_time - sched_dep_time) <= -5)
```
which differs from separating with a comma
```{r filter_5}
filter(flights, (dep_time - sched_dep_time) >= 5, (dep_time - sched_dep_time) <= -5)
```
which returns zero rows (no flight can leave both 5 minutes before and 5 minutes
after the scheduled time).
Filter also works to when using functions in the logical statement. We could
find the longest distance flight on May 27th using
```{r filter_6}
filter(flights, month == 5, day == 27, distance == max(distance, na.rm = TRUE))
```
#### Exercises
From R for Data Science Chapter 5
Find all flights that:
1. Arrived 2 or more hours late
2. Flew to DSM or OMA
3. Operated by UA, AA or DL
4. Arrived more than 2 hours late but didn't leave late
5. Delayed in leaving by at least 1 hour but made up 30 hours in flight
### `mutate()`
`mutate` is used to add new variables to a tibble. As with the other verbs
so far, the first argument is the name of the tibble to mutate followed by a
comma separated list of variables and the values to assign to those variables.
The assignment is done in the form `variable_name = value`.
The `flights` table contains information about the flight time and distance
but does not include the speed. We could combine the air time and the distance
to estimate speed. Let's do that:
```{r mutate_1}
flights <- mutate(flights, speed = distance / (air_time / 60))
```
And now if we view flights, we'll see a new variable called speed:
```{r mutate_2}
select(flights, distance, air_time, speed)
```
Multiple variables can be constructed inside a single `mutate()` call:
```{r}
flights <- mutate(flights,
dep_delay = dep_time - sched_dep_time,
arr_delay = arr_time - sched_arr_time)
select(flights, dep_delay, arr_delay)
```
### `summarize()`
`summarize()` takes as the first argument, you guessed it, the data object and
the following arguments are in the same manner as with mutate. While `mutate()`
must returns an $n$ row column, `summarize()` must return a 1 row column that
is a summary measure (e.g., mean, sd, median). If we wanted to get an idea of
the mean and standard deviation of the arrival delays, we would use
```{r summarize_1}
# na.rm = TRUE just means ignore rows where there is a missing value
summarize(flights,
mean_arrival_delay = mean(arr_delay, na.rm = TRUE),
sd_arrival_delay = sd(arr_delay, na.rm = TRUE))
```
The average flight arrives 6.9 minutes late with a standard deviation of 44.6
minutes.
### `arrange()`
`arrange()` is used to order data by a variable in the data. For instance,
if we wanted to order the `flights` table by the the departure time, we would
run
```{r arrange_1}
arrange(flights, dep_time)
```
and we get a bunch of flights that left at 00:01.
### `group_by()`
The major value of `dplyr` comes with the `group_by` function when combined with
the prior verbs. `group_by` modifies the prior verbs so that they operate on a
group basis. For instance, if we wanted the mean arrival delay by airline,
```{r group_by_1}
flights <- group_by(flights, carrier)
```
You'll notice that `flights` doesn't look much different if you print it
```{r group_by_2}
flights
```
but in the console you'll see the new line that says:
```
# Groups: carrier [16]
```
this indicates that `flights` is now a grouped tibble and the groups are
given by the value of `carrier`. If we apply one of the earlier functions
to `flights` now, such as the summarize function, we'll get a value returned
for each of the groups.
```{r group_by_3}
summarize(flights, mean_arrival_delay = mean(arr_delay, na.rm = TRUE))
```
We can also group by several variables. For instance, maybe flights to one
airport are more delayed than others and there is also a carrier effect. We
would want to group both by carrier and by destination airport:
```{r group_by_4}
flights <- group_by(flights, carrier, dest)
summarize(flights, mean_arrival_delay = mean(arr_delay, na.rm = TRUE))
```
Earlier in the `filter()` examples, we used `filter()` to find the furthest
distance a flight covered. We could do the same trick with `filter()` and
`group_by()` to find the longest-distance flight on each carrier.
```{r group_by_5}
flights <- group_by(flights, carrier)
filter(flights, distance == max(distance))
```
Why is this 12,793 rows and not 16? Multiple flights for the same carrier have
the same distance (e.g., the same route appears several times).
## The Pipe Operator `%>%`
This example is motivated/adapted from R for Data Science.
Imagine the following nursery rhythm:
Little bunny Foo Foo
Went hopping through the forest
Scooping up the field mice
And bopping them on the head
We could code this as:
```
foo_foo <- little_bunny()
foo_foo_1 <- hop(foo_foo, through = forest)
foo_foo_2 <- scoop(foo_foo_1, up = field_mice)
foo_foo_3 <- bop(foo_foo_2, on = head)
```
but that is hard to debug, write and understand. The intermediate steps don't
have clear value, they clutter your workspace and invite error with making
the suffix unique.
You could re-write it so that you simply overwrite the original object in
each step:
```
foo_foo <- little_bunny()
foo_foo <- hop(foo_foo, through = forest)
foo_foo <- scoop(foo_foo, up = field_mice)
foo_foo <- bop(foo_foo, on = head)
```
but this is still hard to read and debug. How do you know when something went
wrong with `foo_foo` is the output is just something is wrong with `foo_foo`?
You could do a lot of nesting of function calls:
```
foo_foo <- little_bunny()
bop(
scoop(
hop(foo_foo, through = forest),
up = field_mice
),
on = head
)
```
but this is confusing because the argument `on` belongs to `bop()` but is
5 lines from the opening of `bop()`.
This rapidly becomes a problem - imagine a typical data summarization step that
involves filtering the data, making a transformation, grouping the data,
making the summary measure and arranging by some value. You'd end up with
```{r pipe_1}
arrange(
summarize(
group_by(
mutate(
filter(flights, origin == "JFK"),
speed = distance / (air_time / 60)),
carrier),
mean_speed = mean(speed, na.rm = TRUE)),
carrier, mean_speed)
```
And that makes about zero sense. It runs but it is confusing to code and
confusing to understand. Instead, it would be amazing to do this naturally with
a "then" type ordering. This is what the `%>%` (pipe) does.
Simply put a pipe takes the results of a function and uses them as the first
argument of the following function. We could re-write the story about Foo Foo
using pipes as
```
foo_foo %>%
hop(through = forest) %>%
scoop(up = field_mouse) %>%
bop(on = head)
```
This is a logical ordering, easy to understand, keeps functions and their
arguments tightly linked and follows naturally.
The data summarization example above can be recast simply as
```{r pipe_2}
flights %>%
filter(origin == "JFK") %>%
mutate(speed = distance / (air_time / 60)) %>%
group_by(carrier) %>%
summarize(mean_speed = mean(speed, na.rm = TRUE)) %>%
arrange(carrier, mean_speed)
```
It gives the same results but is much easier to understand. Since nearly all
data analysis and manipulations are a linear flow from unstructured/messy data
to rectangular data to tidy data to transformations to models, the concept of
"then" as a glue between the steps comes naturally.
## Joins
Typically, data exists in several tables and we would like to merge the data
together into a single table for our analysis. `dplyr` supports a number of
joins. They are:
* `inner_join(x, y)` returns the rows from that exist in both `x` and `y` with
all the values from `x` and `y`
* `left_join(x, y)` returns all the rows in that exist in `x` with the
values from `x` and `y`. If rows exist in `x` but don't have a match in
`y` have their values set to missing (`NA`)
* `right_join(x, y)` is the same as `left_join()` but keeps all the rows in
`y` instead of `x`
* `full_join(x, y)` keeps all the rows and values that occur in either `x` or
`y`
* `semi_join(x, y)` returns all the rows in `x` that have a matching row in `y`
but does not include any variables from `y`
* `anti_join(x, y)` returns all the rows in `x` that do not have a matching row
in `y`
By default, `*_join()` will use all the variables common to `x` and `y` when
performing the join. This behavior can be over-ridden by setting the `by`
argument, see the Arguments section for `inner_join()` for details on how to
do this (hint: `?inner_join`).
We might want to take the data from the `flights` table and merge in the
information about the type and number of engines for each plane. The `flights`
table contains the `tailnum` of the plane that made the flight. The `planes`
table contains more information about that plane. We want to join `flights`
and `planes`, compute average speed and then group that by engine type and
the number of engines and calculate the mean.
```{r joins_1}
mean_speed_by_engine_type_and_number <- flights %>%
ungroup() %>%
select(tailnum, air_time, distance) %>%
inner_join(planes) %>%
select(air_time, distance, engines, engine) %>%
group_by(engine_type = engine, engine_number = engines) %>%
summarize(mean_speed = mean(distance / (air_time / 60),
na.rm = TRUE))
mean_speed_by_engine_type_and_number
```
#### Exercises
How might you find the average and median arrival delay by carrier?
# Visualization
For more information see
[Chapter 3 of R for Data Science](http://r4ds.had.co.nz/data-visualisation.html)
We are going to focus on using `ggplot2` for our plots. The basic use is to
call the function `ggplot(data, mapping = aes())` where data is some
data object (e.g., tibble) and the mapping contains some of the aesthetics
for the plot (e.g., what values should be plotted along the x and y, is there
some grouping in the data that you want to show with color or line or point
type).
After initializing the `ggplot` object with that call, you append `geom`s to
the plot with the `+` operator. For instance, the `geom_point()` produces a
scatter plot of the data set to `x` and `y` in the `mapping` of the
call to `ggplot`. As an example,
```{r vis_1}
ggplot(flights, aes(x = dep_delay, y = arr_delay)) +
geom_point()
```
We might want to color the data points by the carrier
```{r vis_2}
ggplot(flights, aes(x = dep_delay, y = arr_delay, color = carrier)) +
geom_point()
```
We can even combine plots with data manipulations connected through pipes:
```{r vis_3}
flights %>%
inner_join(weather) %>%
ggplot(aes(x = wind_dir,
y = dep_delay,
color = origin)) +
geom_point()
```
This is an interesting plot but hard to understand since the data from the
3 airports are plotted on top of each other. We might want to break those
apart by using what is called a facet or `facet_wrap()` in our plot building
command. `facet_wrap()` takes as its first argument a one-sided formula that
tells it which variable to use to construct the facets. For example,
```{r vis_3b}
flights %>%
inner_join(weather) %>%
ggplot(aes(x = wind_dir,
y = dep_delay,
color = origin)) +
geom_point() +
facet_wrap(~ origin)
```
We can also do things like putting in a smoother to show with the data:
```{r vis_4}
flights %>%
inner_join(weather) %>%
ggplot(aes(x = wind_dir,
y = dep_delay,
color = origin)) +
geom_smooth() +
facet_wrap(~ origin)
```
We didn't pass the argument `method` in `geom_smooth()` and so it defaulted to
using a GAM smoother which is a locally-weighted smoother that performs well
and fast on large data sets. For small data sets, it defaults to `loess` which
is a similar idea as GAM but slower. You can also tell `geom_smooth()` what
type of smoother to use (e.g., `lm` for linear model for a line-of-best-fit).
You might think that the data is describing a circle - 360 and 0 degrees are
the same direction - and wouldn't it be nice if the data was shown with
the x variable forming a circle? We can convert the data to be shown in a
polar coordinate system instead of an Cartesian system by adding `coord_polar()`
to the statement that builds the model:
```{r vis_5}
flights %>%
inner_join(weather) %>%
group_by(origin, wind_dir = round(wind_dir)) %>%
summarize(dep_delay = mean(dep_delay, na.rm = TRUE)) %>%
ggplot(aes(x = wind_dir,
y = dep_delay,
color = origin)) +
geom_point() +
geom_path() +
facet_wrap(~ origin) +
coord_polar()
```
We can take the same graph and improve the labeling or change the theme as
well. The `labs()` argument is used to set the labels for the axis of the plot
while there are a number of themes built-in such as `theme_bw()` or
`theme_minimal()`. Custom themes can be built with the `theme()` function and
the package [`ggthemes`](https://cran.r-project.org/web/packages/ggthemes/vignettes/ggthemes.html)
provides a variety of themes you might want to use.
```{r vis_6}
flights %>%
inner_join(weather) %>%
group_by(origin, wind_dir = round(wind_dir)) %>%
summarize(dep_delay = mean(dep_delay, na.rm = TRUE)) %>%
ggplot(aes(x = wind_dir,
y = dep_delay,
color = origin)) +
geom_point() +
geom_path() +
facet_wrap(~ origin) +
coord_polar() +
labs(x = "Wind Direction", y = "Depature Delay")
```
You can even generate maps in `ggplot2` but this requires that you install
the package `mapproj` as well.
```{r vis_7}
ggplot(airports, aes(x = lon, y = lat)) +
geom_point(size = 0.5) +
borders()
```
# Regression
For more information, see the
[Model Building Chapter in R for Data Science](http://r4ds.had.co.nz/model-building.html)
Regression is the main way in which we estimate the effect of some variable(s)
$X$ on some outcome $y$. You've probably heard of linear regression (line of
best fit). There are other methods known as generalized linear models (GLM)
that are commonly used regression methods as well. These are used when the
response variable has special properties - such as being binary (yes/no,
disease/no disease) or counts. Aaron will talk about logistic regression
next week which is a case of GLM and he will use the `glm()` function.
The syntax does not vary according to the desired model in `glm()`, you just
change the desired family.
In R, a model is estimated by selecting the function we will use to do the
estimation (in this case `glm()`) and then providing a formula that describes
the model. Typically, inside a call to `glm()` you will use 2 or 3
arguments. They are
* formula (first) - the specification of the model you want to estimate. A
formula is written in the form `<response> ~ <x>` in the most
simple case. A more complicated model that includes several possible predictors
is written as `<response> ~ <x1> + <x2> + ... + <xk>`. You can include
transformations in the formula (e.g., `log` or `sqrt`). R will process the
formula and build the model matrix (e.g., it handles construction of dummy
variables for you).
* family (second) - the distribution of the response data. You'll probably
either leave this as the default `gaussian()` for classic linear models or
`binomial()` for binary response data.
* data (third) - the data set to use to estimate the model
For example, we might want to know how the arrival delay is affected by the
departure delay. We would regress the arrival delay in `flights` on the
departure delay to decompose the arrival delay into the portion that is caused
by departure delays and the portion caused by other factors. The data set
we would want to use is `flights`, the model would be `arr_delay ~ dep_delay`
and we can leave the family as `guassian()`. The model building step would
look like:
```{r regression_1}
model <- glm(arr_delay ~ dep_delay,
data = flights)
```
If we want to see a summary of the model fit with hypothesis tests, we would use
the `summary()` function (`summary(x)` applied to nearly any object in R will
produce a summary of `x` that varies based on what `x` is. In this case,
`x` is a model and so it provides a summary of the model fit but if `x` was a
data.frame/tibble, it would provide a variable-by-variable summary including
the min, 1st, 2nd, 3rd quartiles, mean and max.).
```{r regression_2}
summary(model)
```
The `summary(model)` says that a flight that leaves on time arrives about 6
minutes earlier that scheduled and that for each minute that the flight is
delayed compared to the scheduled departure time there is a 1.02 minute delay
on arrival.
# Functional Programming (Probably Won't Get to)
For more detail see [R for Data Science Chapter 25](http://r4ds.had.co.nz/many-models.html).
One of the most powerful things about the tidyverse is the ability do construct
"list-columns" inside a tibble. There look like and behave like normal columns
but instead of containing an atomic value, they contain a tibble. You construct
these with the `nest()` command and the nesting is done by the groups of the
tibble. For instance, if I wanted to nest the data by airline and airport,
I would
```{r nest_1}
nested_flights <- flights %>%
group_by(carrier, origin) %>%
nest()
nested_flights
```
Notice how `carrier` and `origin` are both columns of vectors but the new
variable `data` is a tibble. That tibble contains only the rows of data for that
row's `carrier` and `origin` values.
You can interact with that data using the `purrr` package. For instance, if
we wanted to fit a model that predicted the probability of a plane leaving
late based on the number of flights out, the month, day-of-week, wind speed,
hour and flight distance with a different model for each airline and airport
we could do something like this:
```{r nest_2}
weather_and_time <- flights %>%
ungroup() %>%
inner_join(weather) %>%
select(carrier, origin,
dep_delay,
year, month, day, hour,
wind_speed, distance)
number_of_flights <- flights %>%
group_by(origin, year, month, day, hour) %>%
summarize(number_of_flights_hour = n())
model_data <- inner_join(weather_and_time, number_of_flights) %>%
mutate(day_of_week = as.factor(lubridate::wday(lubridate::ymd(paste(year, month, day)))),
month = as.factor(month)) %>%
select(-year, -day)
model_data
model_data <- model_data %>%
group_by(carrier, origin) %>%
nest() %>%
filter(carrier %in% c("AA", "UA", "DL"))
model_data
model_data <- model_data %>%
mutate(model = purrr::map(data, ~ lm(dep_delay ~ ., data = .)))
model_data
```
We now have a column named `model` that contains a series of 35 linear models.
We can see that these are models if we select the vector and print the first
element:
```{r}
model_data$model[[1]]
```
We can use the `broom` package to extract the estimated coefficients of the
model and see if the different airlines have different sensitivities to the
number of flights out of the airport:
```{r}
sensitivity_to_congestion <- model_data %>%
mutate(sens_to_congestion = purrr::map(model,
~ broom::tidy(.) %>%