diff --git a/README.md b/README.md
index d47f928..2c73897 100644
--- a/README.md
+++ b/README.md
@@ -7,6 +7,13 @@
3.3. Output
3.4. Example dataset
4. Visualization
+4.1. Getting started
+4.2. Genome browser layout
+4.3. Free layout mode
+4.4. Customizing visualization
+4.5. Highlighting paths
+4.6. Understanding visualization
+4.7. Navigation
5. RNA-Seq scaffolder
6. References
7. Feedback and bug reports
@@ -141,7 +148,7 @@ Note that we assume that SGTK installation directory is added to the `PATH` vari
-## Command line options
+## 3.1. Command line options
All input options are capable of taking only a single or a pair of files if specified. To provide multiple files use the same option again (e.g. `-c contigs1.fa -c contigs2.fa`). See more in the examples.
`-h` (or `--help`)
@@ -286,7 +293,7 @@ For example:
e 1 3 1 0 32.5 200
-## Command line examples
+## 3.2. Command line examples
Let's say our dataset consists of:
- A set of contigs (`contigs.fa`)
@@ -309,7 +316,7 @@ Then the command line for launching SGTK would look like:
-o output_dir
-## Output
+## 3.3. Output
SGTK stores all output files in ` `, which is set by the user.
@@ -319,7 +326,7 @@ SGTK stores all output files in ` `, which is set by the user.
Open `main.html` in any browser to see visualisation. Chrome is preferred.
-## Example dataset
+## 3.4. Example dataset
SGTK comes with toy dataset, on which you can test your installation:
@@ -358,7 +365,10 @@ This example is constructed from E.coli paired-end reads and pacbio reads. Conti
# 4. Visualization
-## Getting started
+
+
+## 4.1. Getting started
+
After the graph is constructed and the web page is generated, you can open `main.html` in a web browser (we recommended to use Chrome, however it also was tested in FireFox, Opera and Safari).
@@ -369,14 +379,15 @@ SGTK has two layout modes: (i) free layout and (ii) genome browser. Free layout
-## Genome browser layout
+
+## 4.2. Genome browser layout
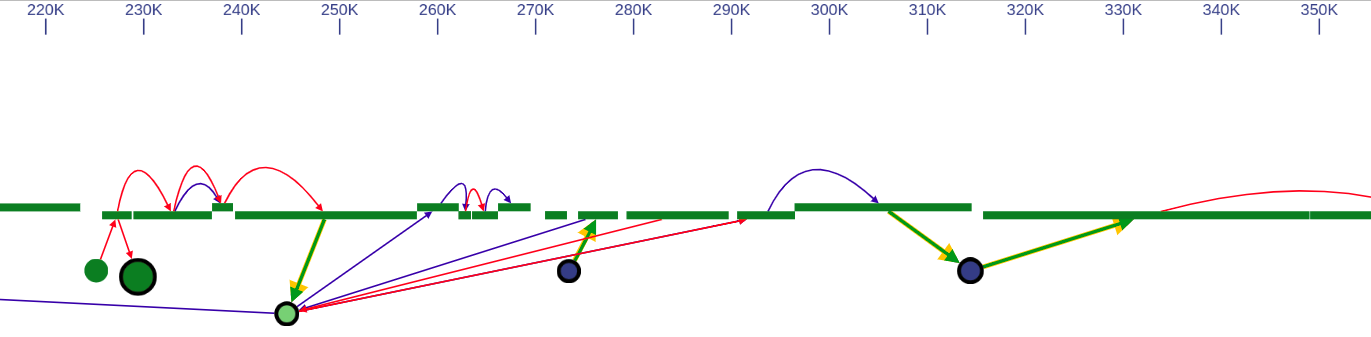
In the genome browser mode, vertices of the scaffold graph are displayed as rectangles placed along the reference, lengths of which are proportional to the contigs sizes. Short nodes with adjacent edges are hidden and rendered only when zoomed in.
-
-## Free layout mode
+
+## 4.3. Free layout mode
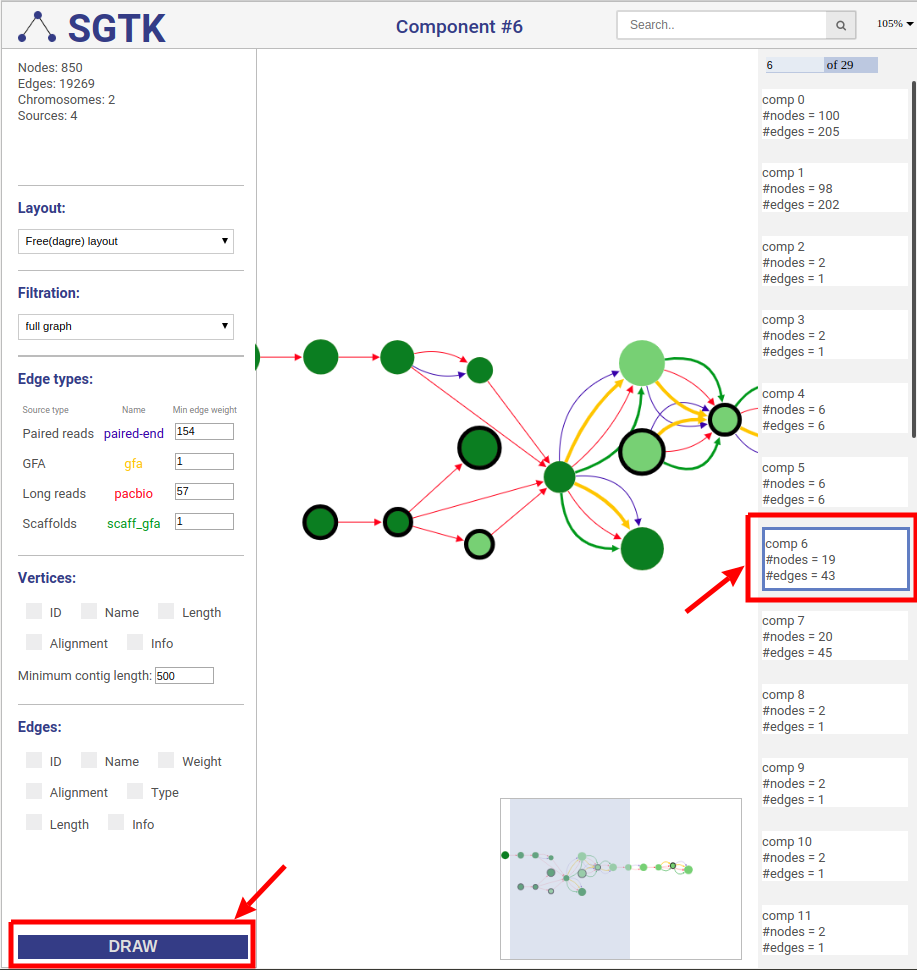
In case of free layout you can choose one of several filtration options. Once new visualization parameters are set, click the `DRAW` button and choose the component.
@@ -434,7 +445,8 @@ Visualizes the local area of the specified size for vertices that have more than
-## Customizing visualization
+
+## 4.4. Customizing visualization
### Connection sources
@@ -470,64 +482,76 @@ Using controls displayed below you can set which properties will be shown near v
You can set up threshold for visualizing nodes. Note, that when visualizing nodes along scaffolds contigs shorter than the threshold still will be displayed.
-## Path highlight
+
+## 4.5. Highlighting paths
+
+To ease the analysis, you can also highlight vertices, edges, paths and entire scaffolds.
+To use this functionality, switch to highlight tab (top left).
+
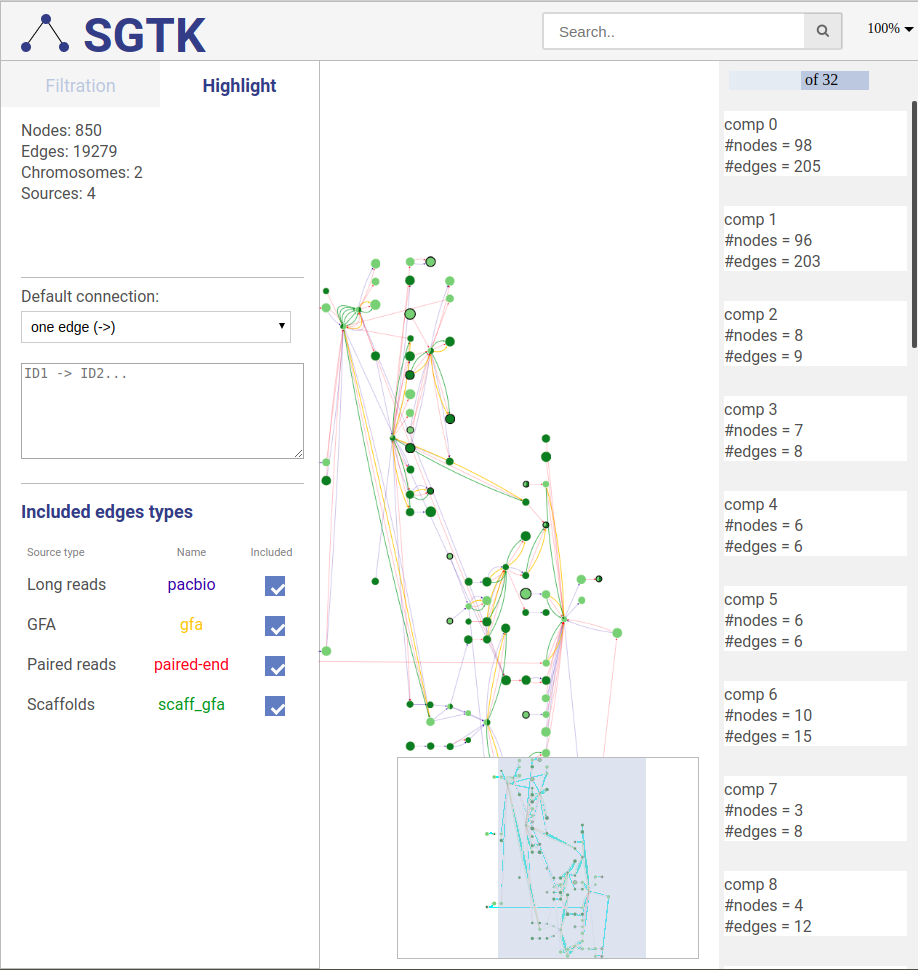
-You can highlight vertex, edges, pathes and scaffolds.
-Open highlight pan for doing that.
### Highlighted path description
#### Path description
-
+To highlight a set of edges or vertices, one needs to specify them in the text box using simple description script.
-You can write highlight description to that window.
+There are following types of highlight description are currently supported:
+* `scaffold_name` - scaffold name to highlight; will highlight all vertices and connections corresponding to this scaffold.
+* `a` - contig name or contig id to highlight the corresponding vertex.
+* `a --> b` - will find and highlight the shortest path between vertices a and b.
+* `a -> b` - highlight vertices a and b and all connections between them.
+* `a; b` - highlight vertices a and b without highlighting any connections between them.
+* `a -eid> b` - highlight vertices a and b, as well as edge eid if it connects these two vertices.
+* `a b` in this case the default connection type between vertices a and b will be used (see below).
-There are following types of description:
-* `scaffold_name` - scaffold name for highlight all vertex and connections corresponding to that scaffold.
-* `a` - contig name or contig id to highlight vertex.
-* `a --> b` - will be find and highlight the shortest path between vertices.
-* `a -> b` - highlight contigs a and b and connections between them.
-* `a; b` - highlight a and b indipendatly.
-* `a -eid> b` - highlight contigs a, b and edge with eid if it connected this two vertices.
-* `a b` in this case will be used default connection between contigs a and b.
+Each vertex is specified by corresponding contig name or id.
-a and b are contigs name or id.
+
-#### Tapholding
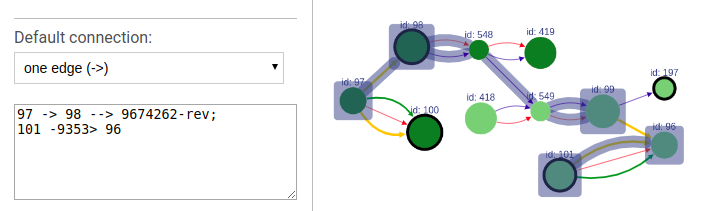
+#### Default connection
-You can add vertex to the end of highlighting list by tapholding the vertex.
+To choose the default connection type use the appropriate drop-down menu. Available types are: path, edge or none.
-#### Default connection
-You can choose which connection will be use by default: path, edge or none.
-### Fake edges
+#### Extending highlighted paths
-
+To extend currently highlighted path, you can hold your left mouse button while pointing on the vertex of interest.
-In case of edge or path absent fake connection between
-two vertices will be red highlighting.
-### Path extension
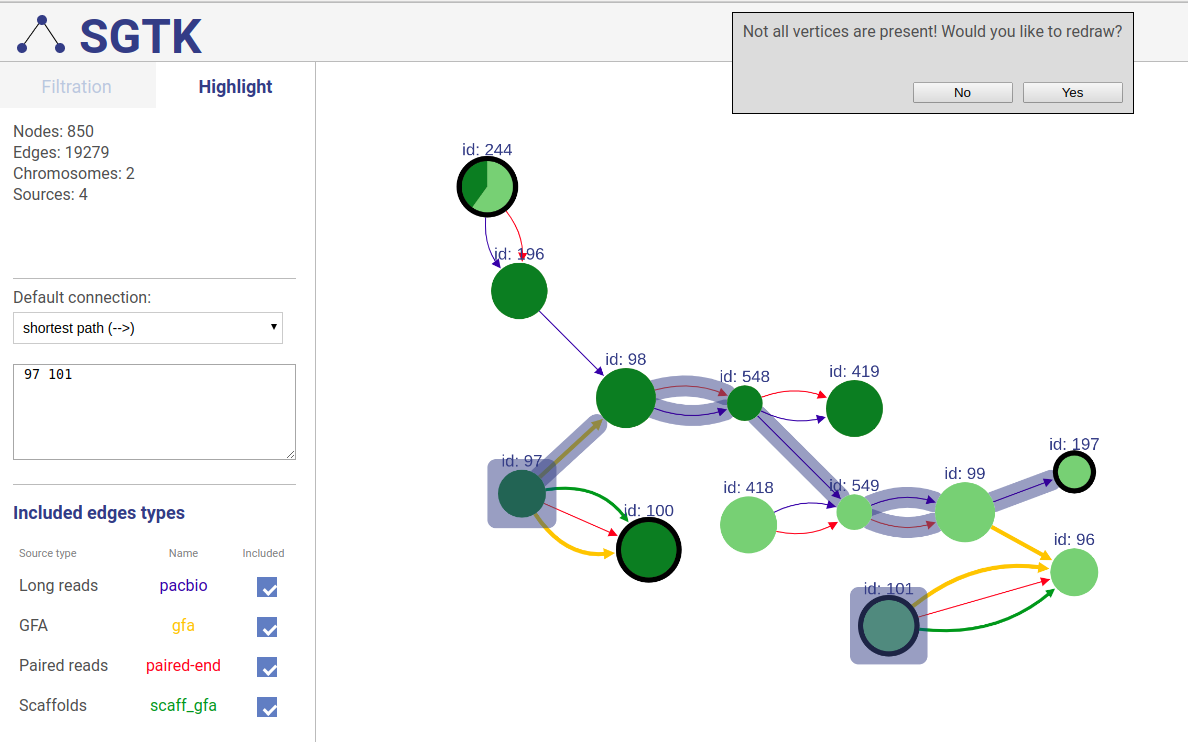
+### Extending displayed component
+If a path or a set of vertices/edges chosen to be highlighted is not entirely displayed in the current view, you will be prompted about extending the view and showing absent vertices.
+
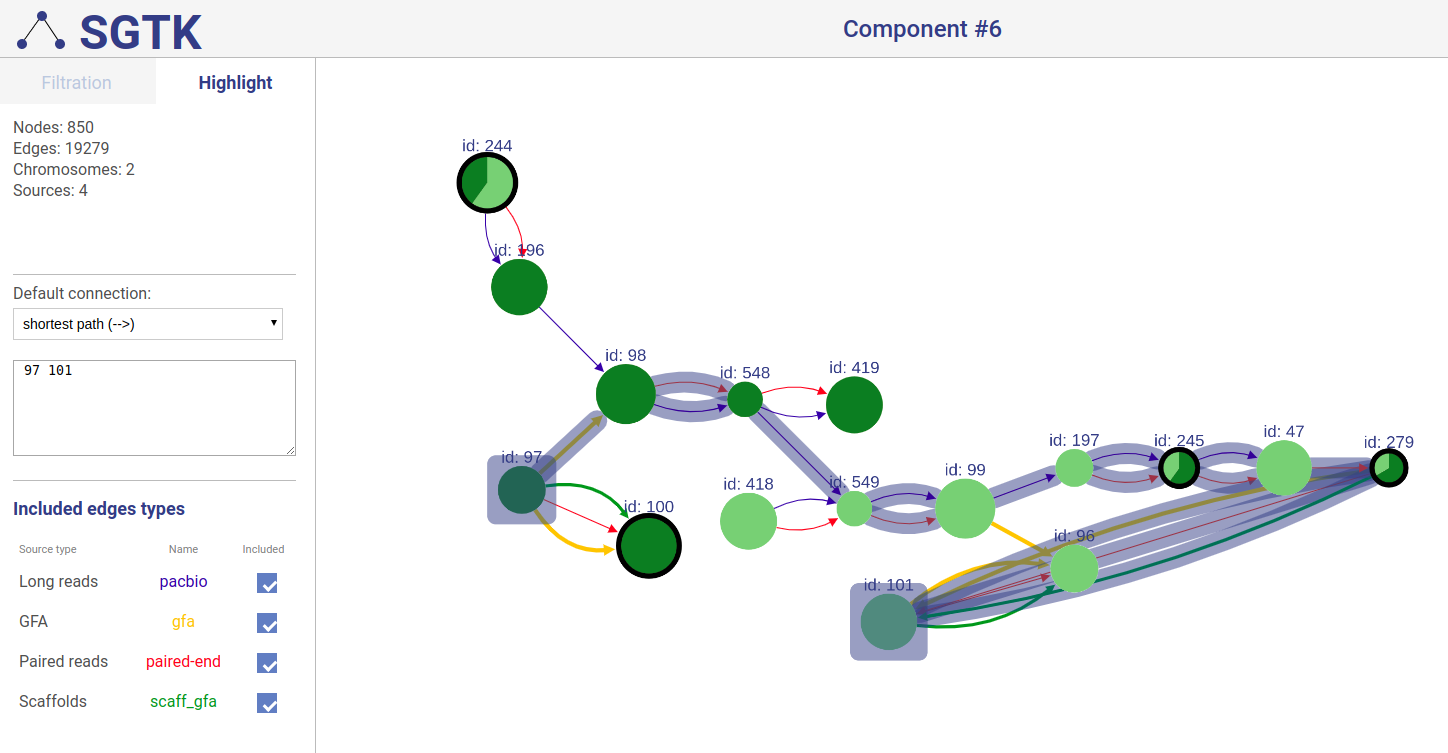
-If the path not completely present on screen
-you will be asked about showing absent vertices.
-### Included sources
+
+### Included connection sources
+
+You can also choose which connection types will be highlighted.
+
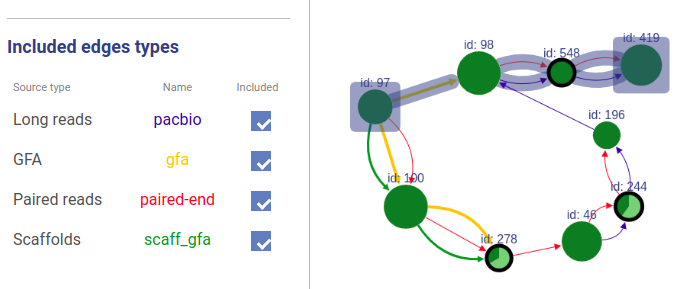
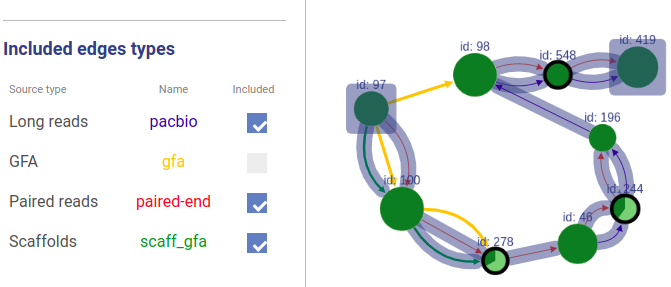
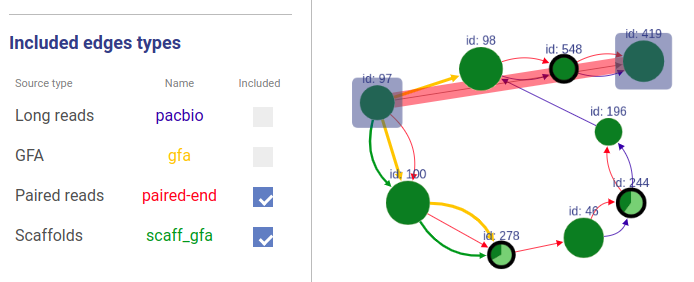
-You can choose which sources to take into account in path highlighting.
-## Understanding visualization
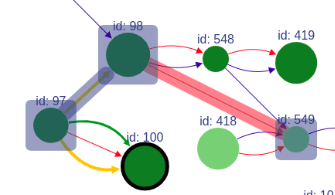
+### Missing edges
+
+If a user asks to highlight an edge between two vertices that are not connected, a fake edge will be drawn between them and highlighted with red color (instead of typical blue highlight).
+Such option can be useful, for example, when comparing scaffolds obtained with different methods.
+
+
+
+
+
+
+## 4.6. Understanding visualization
Below we provide brief information on how interprete SGTK visualization.
@@ -591,7 +615,8 @@ When the courson is pointed over a graph element, information about this node or
-## Navigation
+
+## 4.7. Navigation
### View navigator