MetaDegron: Multimodal feature-integrated protein language model for predicting E3 ligase targeted degrons.
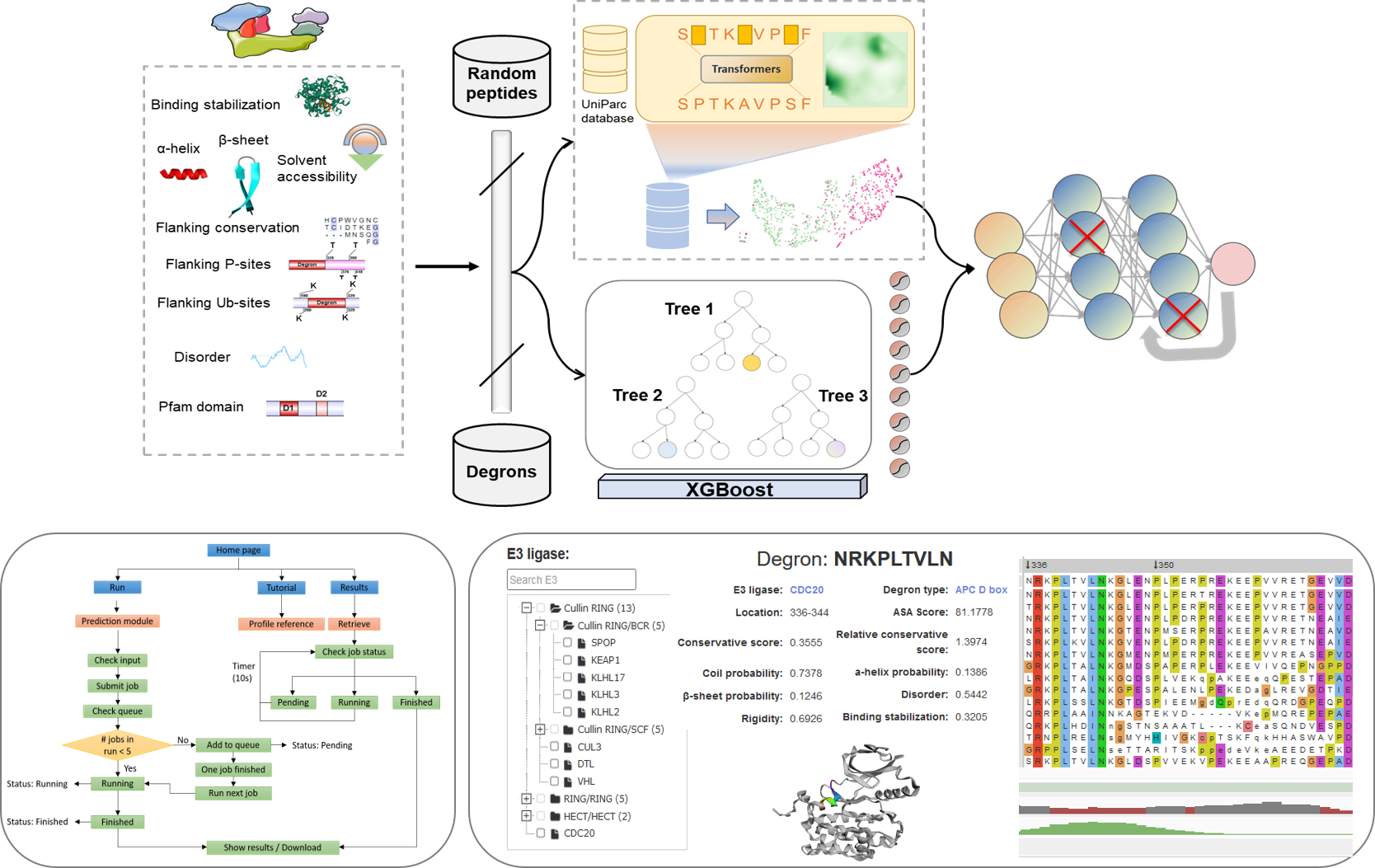
Protein degradation via the ubiquitin proteasome system (UPS) at the spatial and temporal regulation is essential for many cellular processes. E3 ligases and degrons, the sequences they recognize in the target proteins, are key parts of the ubiquitin-mediated proteolysis, and their interactions determine the degradation specificity and maintain cellular homeostasis. So far, only a small number of targeted degron instances have been identified, and their properties remain to be systematically characterized. To tackle on this great challenge, here we develop a novel deep-learning framework, namely MetaDegron for predicting E3 ligase targeted degron by integrating the protein language model and comprehensive featurization strategies. Through extensive evaluation on benchmark datasets and comparison with state-of-the-art method, we demonstrate the superior performance of MetaDegron in terms of prediction accuracy. Among functional features, MetaDegron allows batch prediction of targeted degrons of 21 E3 ligases, and provides functional annotations and visualization of multiple degron-related structural and physicochemical features. MetaDegron is freely available at http://modinfor.com/MetaDegron/. We anticipate MetaDegron can serve as a useful tool for the clinical and translational community to elucidate the mechanisms of regulation of protein homeostasis, cancer research, and drug development.
Download MetaDegron by
git clone https://github.com/BioDataStudy/MetaDegron.git
Installation has been tested in Linux server, CentOS Linux release 7.8.2003 (Core), with Python 3.7. Since the package is written in python 3x, python3x with the pip tool must be installed. MetaDegron uses the following dependencies: numpy, scipy, pandas, h5py, torch, allennlp, keras version=2.3.1, tensorflow=1.15 shutil, and pathlib. We highly recommend that users leave a message under the MetaDegron issue interface (https://github.com/BioDataStudy/MetaDegron/issue) when encountering any installation and running problems. We will deal with it in time. You can install these packages by the following commands:
conda create -n MetaDegron python=3.7
conda activate MetaDegron
pip install pandas
pip install numpy
pip install scipy
pip install torch
pip install allennlp==0.9.0
pip install -v keras==2.3.1
pip install -v tensorflow==1.15
pip install seaborn
pip install shutil
pip install protobuf==3.20
pip install h5py==2.10.0
Please download large protein language model from the http://modinfor.com/MetaDegron/Download/weights.hdf5. Note: right click - > save the link
Copy or move the weights.hdf5 file to the models\uniref50_v2\ directory to ensure that the model is preloaded successfully
Please download the training dataset (>200M, Unable to upload to github) from the http://modinfor.com/MetaDegron/Download/all_train_embedding.npy. Note: right click - > save the link
Copy or move the all_train_embedding.npy file to the data/ directory to ensure that the train_embedding can be loaded successfully
Researchers can run MetaDegron online at http://modinfor.com/MetaDegron/. For commercial usage inquiries, please contact the authors.
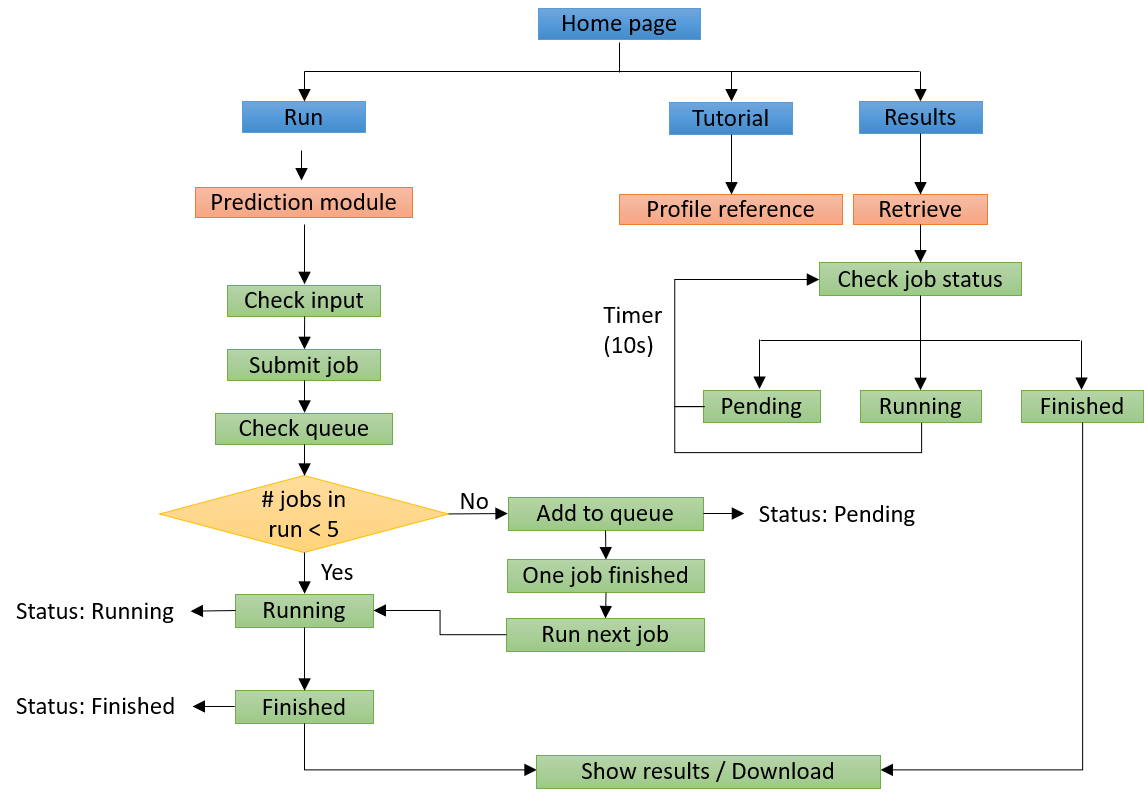
The workflow of the MetaDegron online server is illustrated below. The information given is initially checked by the web server for proper formatting, among other things. MetaDegron launches a new job and changes the state of the work to "Running" after the input data is checked for correct format. Otherwise, MetaDegron moves the new work to the bottom of the queue and changes its status to "Pending." Users are then routed to a website for tracking the job status after a job is successfully submitted, where the status is updated every 10 seconds until the task is complete. Through the unique job identifier, which can be generated automatically or customized by the submitter, users are able to monitor the job status and retrieve the result.
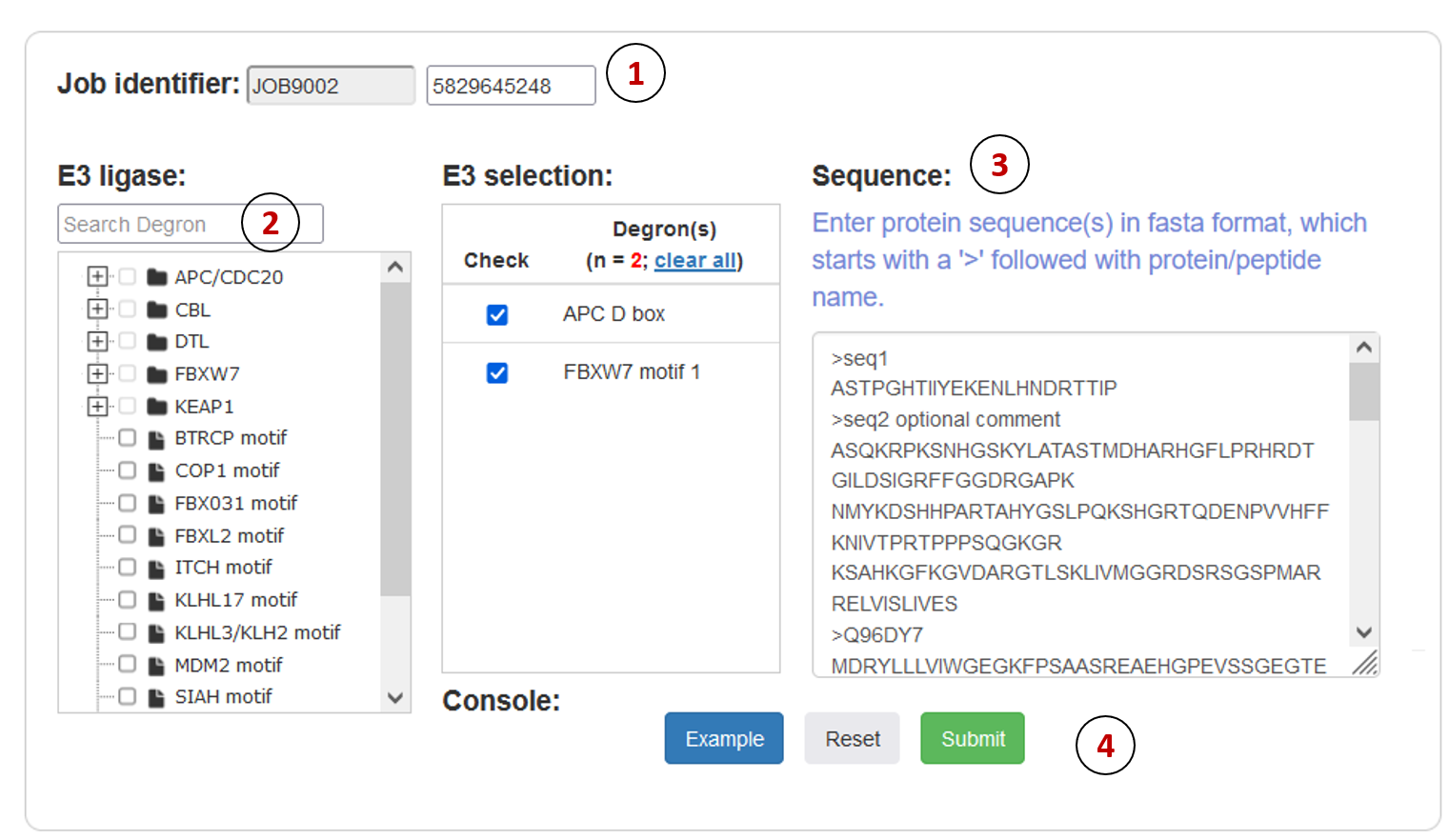
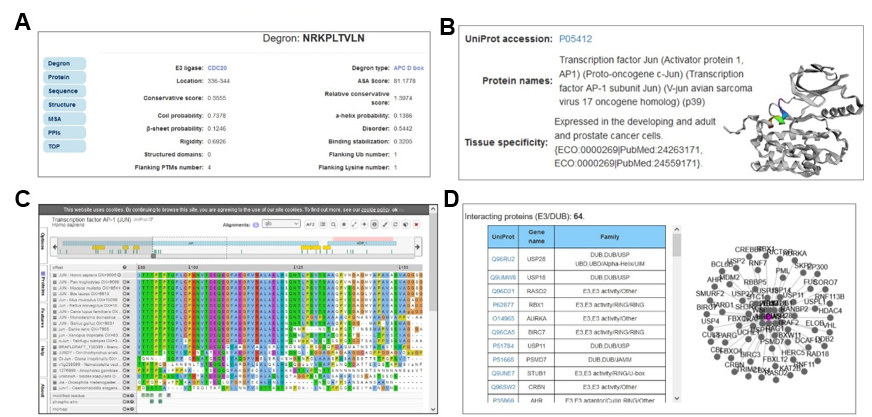
MetaDegron serves as a useful tool for predicting targeted degrons of 21 E3 ligases, offering researchers possible candidates for studying protein degradation pathways and identifying potential therapeutic targets. The multimodal feature integration approach enables MetaDegron to capture diverse aspects of degron recognition, including amino acid composition, physicochemical properties, evolutionary conservation, and contextual dependencies, thereby enhancing its capabilities for advancing research in the field of protein degradation and ubiquitin-mediated proteolysis. The webserver of MetaDegron was designed and constructed with a modular and user-friendly manner. Three major modules, including “Run”, “Results” and “Tutorial”, are the kernel of MetaDegron online server. The ‘Run’ module sequentially controls the execution of submitted jobs, including the input checking, job submitting, job running, and task terminates. Meanwhile, the “Results” module records the submission jobs, monitors the status of jobs, and immediately shows the prediction results. The clickable and searchable hierarchical classification tree of E3s is loaded for the selection of single or multiple E3 ligases. Then, one or more protein sequences in FASTA format can be submitted. After finishing the submitted job, the prediction results will be visualized with specific information, including the “Entry”, “E3 ligase”, “Degron instance”, “Degron type”, “Start”, “End”, and “Score”. It displays the detailed information for degron and source protein
- Job identifier: Job identifier can be generated automatically or customized by the submitter. It is confidential to other users and can be used for job status monitoring and result retrieval.(See Results page).It is required.
- E3 ligase: The MetaDegron 1.0 server supports 21 E3 ligase prediction. We constructed a classification tree of E3 ligase. Users can quickly retrieve and submit candidate E3 through the search box and tree map.
- Sequence: User can directly copy one or more proteins with FASTA format in the input box.
- Operation buttons: Submit, reset the submission form, or access the example dataset.
Please cite the following paper for using: Multimodal feature-integrated protein language model for predicting E3 ligase targeted degrons. In submission.