Author: Daniel Steinberg
License: LGPL v3 (See COPYING and COPYING.LESSER)
Overview:
This library implements the following algorithms with variational Bayes learning procedures and efficient cluster splitting heuristics:
- The Variational Dirichlet Process (VDP) [1, 2, 6]
- The Bayesian Gaussian Mixture Model [3 - 6]
- The Grouped Mixtures Clustering (GMC) model [6]
- The Symmetric Grouped Mixtures Clustering (S-GMC) model [4 - 6]. This is referred to as Gaussian latent Dirichlet allocation (G-LDA) in [4, 5].
- Simultaneous Clustering Model (SCM) for Multinomial Documents, and Gaussian Observations [5, 6].
- Multiple-source Clustering Model (MCM) for clustering two observations, one of an image/document, and multiple of segments/words simultaneously [4 - 6].
- And more clustering algorithms based on diagonal Gaussian, and Exponential distributions.
And also,
- Various functions for evaluating means, standard deviations, covariance, primary Eigenvalues etc of data.
- Extensible template interfaces for creating new algorithms within the variational Bayes framework.

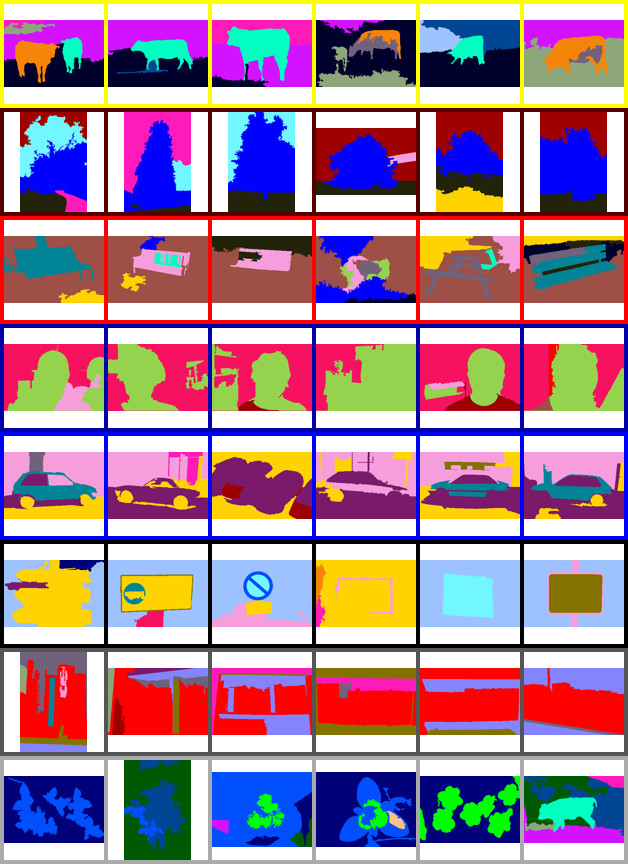
An example of using the MCM to simultaneously cluster images and objects within images for unsupervised scene understanding. See [4 - 6] for more information.
- Eigen version 3.0 or greater
- Boost version 1.4.x or greater and devel packages (special math functions)
- OpenMP, comes default with most compilers (may need a special version of LLVM).
- CMake
For the python interface:
- Python 2 or 3
- Boost python and boost python devel packages (make sure you have version 2 or 3 for the relevant version of python)
- Numpy (tested with v1.7)
For Linux and OS X -- I've never tried to build on Windows.
To build libcluster:
-
Make sure you have CMake installed, and Eigen and Boost preferably in the usual locations:
/usr/local/include/eigen3/ or /usr/include/eigen3 /usr/local/include/boost or /usr/include/boost -
Make a build directory where you checked out the source if it does not already exist, then change into this directory,
cd {where you checked out the source} mkdir build cd build -
To build libcluster, run the following from the build directory:
cmake .. make sudo make installThis installs:
libcluster.h /usr/local/include distributions.h /usr/local/include probutils.h /usr/local/include libcluster.* /usr/local/lib (* this is either .dylib or .so) -
Use the doxyfile in {where you checked out the source}/doc to make the documentation with doxygen:
doxygen Doxyfile
NOTE: There are few options you can change using ccmake (or the cmake gui), these include:
-
BUILD_EXHAUST_SPLIT(toggleONorOFF, defaultOFF) This uses the exhaustive cluster split heuristic [1, 2] instead of the greedy heuristic [4, 5] for all algorithms but the SCM and MCM. The greedy heuristic is MUCH faster, but does give different results. I have yet to determine whether it is actually worse than the exhaustive method (if it is, it is not by much). The SCM and MCM only use the greedy split heuristic at this stage. -
BUILD_PYTHON_INTERFACE(toggleONorOFF, defaultOFF) Build the python interface. This requires boost python, and also uses row-major storage to be compatible with python. -
BUILD_USE_PYTHON3(toggleONorOFF, defaultON) Use python 3 or 2 to build the python interface. Make sure you have the relevant python and boost python libraries installed! -
CMAKE_INSTALL_PREFIX(default/usr/local) The default prefix for installing the library and binaries. -
EIGEN_INCLUDE_DIRS(default/usr/include/eigen3) Where to look for the Eigen matrix library.
NOTE: On linux you may have to run sudo ldconfig before the system can
find libcluster.so (or just reboot).
NOTE: On Red-Hat based systems, /usr/local/lib is not checked unless
added to /etc/ld.so.conf! This may lead to "cannot find libcluster.so"
errors.
All of the interfaces to this library are documented in include/libcluster.h.
There are far too many algorithms to go into here, and I strongly recommend
looking at the test/ directory for example usage, specifically,
cluster_test.cppfor the group mixture models (GMC etc)scluster_test.cppfor the SCMmcluster_test.cppfor the MCM
Here is an example for regular mixture models, such as the BGMM, which simply clusters some test data and prints the resulting posterior parameters to the terminal,
#include "libcluster.h"
#include "distributions.h"
#include "testdata.h"
//
// Namespaces
//
using namespace std;
using namespace Eigen;
using namespace libcluster;
using namespace distributions;
//
// Functions
//
// Main
int main()
{
// Populate test data from testdata.h
MatrixXd Xcat;
vMatrixXd X;
makeXdata(Xcat, X);
// Set up the inputs for the BGMM
Dirichlet weights;
vector<GaussWish> clusters;
MatrixXd qZ;
// Learn the BGMM
double F = learnBGMM(Xcat, qZ, weights, clusters, PRIORVAL, true);
// Print the posterior parameters
cout << endl << "Cluster Weights:" << endl;
cout << weights.Elogweight().exp().transpose() << endl;
cout << endl << "Cluster means:" << endl;
for (vector<GaussWish>::iterator k=clusters.begin(); k < clusters.end(); ++k)
cout << k->getmean() << endl;
cout << endl << "Cluster covariances:" << endl;
for (vector<GaussWish>::iterator k=clusters.begin(); k < clusters.end(); ++k)
cout << k->getcov() << endl << endl;
return 0;
}
Note that distributions.h has also been included. In fact, all of the
algorithms in libcluster.h are just wrappers over a few key functions in
cluster.cpp, scluster.cpp and mcluster.cpp that can take in arbitrary
distributions as inputs, and so more algorithms potentially exist than
enumerated in libcluster.h. If you want to create different algorithms, or
define more cluster distributions (like categorical) have a look at inheriting
the WeightDist and ClusterDist base classes in distributions.h. Depending
on the distributions you use, you may also have to come up with a way to
'split' clusters. Otherwise you can create an algorithm with a random initial
set of clusters like the MCM at the top level, which then variational Bayes
will prune.
There are also some generally useful functions included in probutils.h when
dealing with mixture models (such as the log-sum-exp trick).
Easy, follow the normal build instructions up to step (4) (if you haven't already), then from the build directory:
cmake ..
ccmake .
Make sure BUILD_PYTHON_INTERFACE is ON
make
sudo make install
This installs all the same files as step (4), as well as libclusterpy.so to
your python staging directory, so it should be on your python path. I.e. just
run
import libclusterpyTrouble Shooting:
On Fedora 20/21 I have to append /usr/local/lib to the file /etc/ld.so.conf
to make python find the compiled shared object.
Import the library as
import numpy as np
import libclusterpy as lcThen for the mixture models, assuming X is a numpy array where X.shape is
(N, D) -- N being the number of samples, and D being the dimension of
each sample,
f, qZ, w, mu, cov = lc.learnBGMM(X)
where f is the final free energy value, qZ is a distribution over all of
the cluster labels where qZ.shape is (N, K) and K is the number of
clusters (each row of qZ sums to 1). Then w, mu and cov the expected
posterior cluster parameters (see the documentation for details. Alternatively,
tuning the prior argument can be used to change the number of clusters found,
f, qZ, w, mu, cov = lc.learnBGMM(X, prior=0.1)
This interface is common to all of the simple mixture models (i.e. VDP, BGMM etc).
For the group mixture models (GMC, SGMC etc) X is a list of arrays of size
(Nj, D) (indexed by j), one for each group/album, X = [X_1, X_2, ...]. The
returned qZ and w are also lists of arrays, one for each group, e.g.,
f, qZ, w, mu, cov = lc.learnSGMC(X)
The SCM again has a similar interface to the above models, but now X is a
list of lists of arrays, X = [[X_11, X_12, ...], [X_21, X_22, ...], ...].
This specifically for modelling situations where X is a matrix of all of the
features of, for example, N_ij segments in image ij in album j.
f, qY, qZ, wi, wij, mu, cov = lc.learnSCM(X)
Where qY is a list of arrays of top-level/image cluster probabilities, qZ
is a list of lists of arrays of bottom-level/segment cluster probabilities.
wi are the mixture weights (list of arrays) corresponding to the qY labels,
and wij are the weights (list of lists of arrays) corresponding the qZ
labels. This has two optional prior inputs, and a cluster truncation level
(max number of clusters) for the top-level/image clusters,
f, qY, qZ, wi, wij, mu, cov = lc.learnSCM(X, trunc=10, dirprior=1,
gausprior=0.1)
Where dirprior refers to the top-level cluster prior, and gausprior the
bottom-level.
Finally, the MCM has a similar interface to the MCM, but with an extra input,
W which is of the same format as the X in the GMC-style models, i.e. it is
a list of arrays of top-level or image features, W = [W_1, W_2, ...]. The
usage is,
f, qY, qZ, wi, wij, mu_t, mu_k, cov_t, cov_k = lc.learnMCM(W, X)
Here mu_t and cov_t are the top-level posterior cluster parameters -- these
are both lists of T cluster parameters (T being the number of clusters
found. Similarly mu_k and cov_k are lists of K bottom-level posterior
cluster parameters. Like the SCM, this has a number of optional inputs,
f, qY, qZ, wi, wij, mu_t, mu_k, cov_t, cov_k = lc.learnMCM(W, X, trunc=10,
gausprior_t=1,
gausprior_k=0.1)
Where gausprior_t refers to the top-level cluster prior, and gausprior_k
the bottom-level.
Look at the libclusterpy docstrings for more help on usage, and the
testapi.py script in the python directory for more usage examples.
NOTE if you get the following message when importing libclusterpy:
ImportError: /lib64/libboost_python.so.1.54.0: undefined symbol: PyClass_Type
Make sure you have boost-python3 installed!
When verbose mode is activated you will get output that looks something like this:
Learning MODEL X...
--------<=>
---<==>
--------x<=>
--------------<====>
----<*>
---<>
Finished!
Number of clusters = 4
Free Energy = 41225
What this means:
-iteration of Variational Bayes (VBE and VBM step)<cluster splitting has started (model selection)=found a valid candidate split>chosen candidate split and testing for inclusion into modelxclusters have been deleted because they became devoid of observations*clusters (image/document clusters) that are empty have been removed.
For best clustering results, I have found the following tips may help:
-
If clustering runs REALLY slowly then it may be because of hyper-threading. OpenMP will by default use as many cores available to it as possible, this includes virtual hyper-threading cores. Unfortunately this may result in large slow-downs, so try only allowing these functions to use a number of threads less than or equal to the number of PHYSICAL cores on your machine.
-
Garbage in = garbage out. Make sure your assumptions about the data are reasonable for the type of cluster distribution you use. For instance, if
your observations do not resemble a mixture of Gaussians in feature space, then it may not be appropriate to use Gaussian clusters. -
For Gaussian clusters: standardising or whitening your data may help, i.e.
if X is an NxD matrix of observations you wish to cluster, you may get better results if you use a standardised version of it, X*,
X_s = C * ( X - mean(X) ) / std(X)where
Cis some constant (optional) and the mean and std are for each column of X.You may obtain even better results by using PCA or ZCA whitening on X (assuming ZERO MEAN data), using python syntax:
[U, S, V] = svd(cov(X)) X_w = X.dot(U).dot(diag(1. / sqrt(diag(S)))) # PCA WhiteningSuch that
cov(X_w) = I_D.Also, to get some automatic scaling you can multiply the prior by the PRINCIPAL eigenvector of
cov(X)(orcov(X_s),cov(X_w)).NOTE: If you use diagonal covariance Gaussians I STRONGLY recommend PCA or ZCA whitening your data first, otherwise you may end up with hundreds of clusters!
-
For Exponential clusters: Your observations have to be in the range [0, inf). The clustering solution may also be sensitive to the prior. I find usually using a prior value that has the approximate magnitude of your data or more leads to better convergence.
[1] K. Kurihara, M. Welling, and N. Vlassis. Accelerated variational Dirichlet process mixtures, Advances in Neural Information Processing Systems, vol. 19, p. 761, 2007.
[2] D. M. Steinberg, A. Friedman, O. Pizarro, and S. B. Williams. A Bayesian nonparametric approach to clustering data from underwater robotic surveys. In International Symposium on Robotics Research, Flagstaff, AZ, Aug. 2011.
[3] C. M. Bishop. Pattern Recognition and Machine Learning. Cambridge, UK: Springer Science+Business Media, 2006.
[4] D. M. Steinberg, O. Pizarro, S. B. Williams. Synergistic Clustering of Image and Segment Descriptors for Unsupervised Scene Understanding, In International Conference on Computer Vision (ICCV). IEEE, Sydney, NSW, 2013.
[5] D. M. Steinberg, O. Pizarro, S. B. Williams. Hierarchical Bayesian Models for Unsupervised Scene Understanding. Journal of Computer Vision and Image Understanding (CVIU). Elsevier, 2014.
[6] D. M. Steinberg, An Unsupervised Approach to Modelling Visual Data, PhD Thesis, 2013.
Please consider citing the following if you use this code:
- VDP: [2, 4, 6]
- BGMM: [5, 6]
- GMC: [6]
- SGMC/GLDA: [4, 5, 6]
- SCM: [5, 6]
- MCM: [4, 5, 6]
You can find these on my homepage. Thank you!