-
Notifications
You must be signed in to change notification settings - Fork 39
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
- Loading branch information
Showing
6 changed files
with
152 additions
and
0 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,152 @@ | ||
| # Use peptide prediction models from Koina for MSBooster feature generation | ||
|
|
||
| [Koina](https://koina.proteomicsdb.org/) is a web server that allows users to get MS2, retention time (RT), and ion mobility (IM) | ||
| predictions from multiple machine and deep-learning models from the community. It can be tedious running each model | ||
| separately due to differences in supported peptides, string formatting, and accessibility. Koina combats this by | ||
| providing a central hub where all models can be queried in a similar fashion. | ||
| [MSBooster]([Koina](https://koina.proteomicsdb.org/)) uses the Koina API to gain | ||
| access to these different models, some of which will outperform the FragPipe default of DIA-NN depending on the types of | ||
| data they were trained on. Below we demonstrate two different ways to use Koina, either by explicitly choosing a model | ||
| of your choice or by letting a heuristic best model search algorithm make that decision for you. | ||
|
|
||
| ## Example dataset: HLA | ||
|
|
||
| This tutorial uses one mzML file from [the HLA Ligand Atlas](https://pubmed.ncbi.nlm.nih.gov/33858848/). | ||
| It has been searched by MSFragger, producing a pin file, a pepXML file, and a fragger.params file. All files can be | ||
| downloaded from the [Google Drive link here](https://drive.google.com/drive/folders/1szQmsLD2d_lE3JAWcnRcLR8rcjiGqdHb?usp=sharing). | ||
| 1. If you have not already, please download the [latest FragPipe](https://github.com/Nesvilab/FragPipe/releases). | ||
| Koina is available as of version 22.0. | ||
| 2. Load your mzML file. | ||
|
|
||
|  | ||
|
|
||
| 3. Load the "Nonspecific-HLA" workflow from the workflow tab. Then you can uncheck the MSFragger checkbox, only running | ||
| the steps in the validation tab. It should look like this. | ||
|
|
||
| 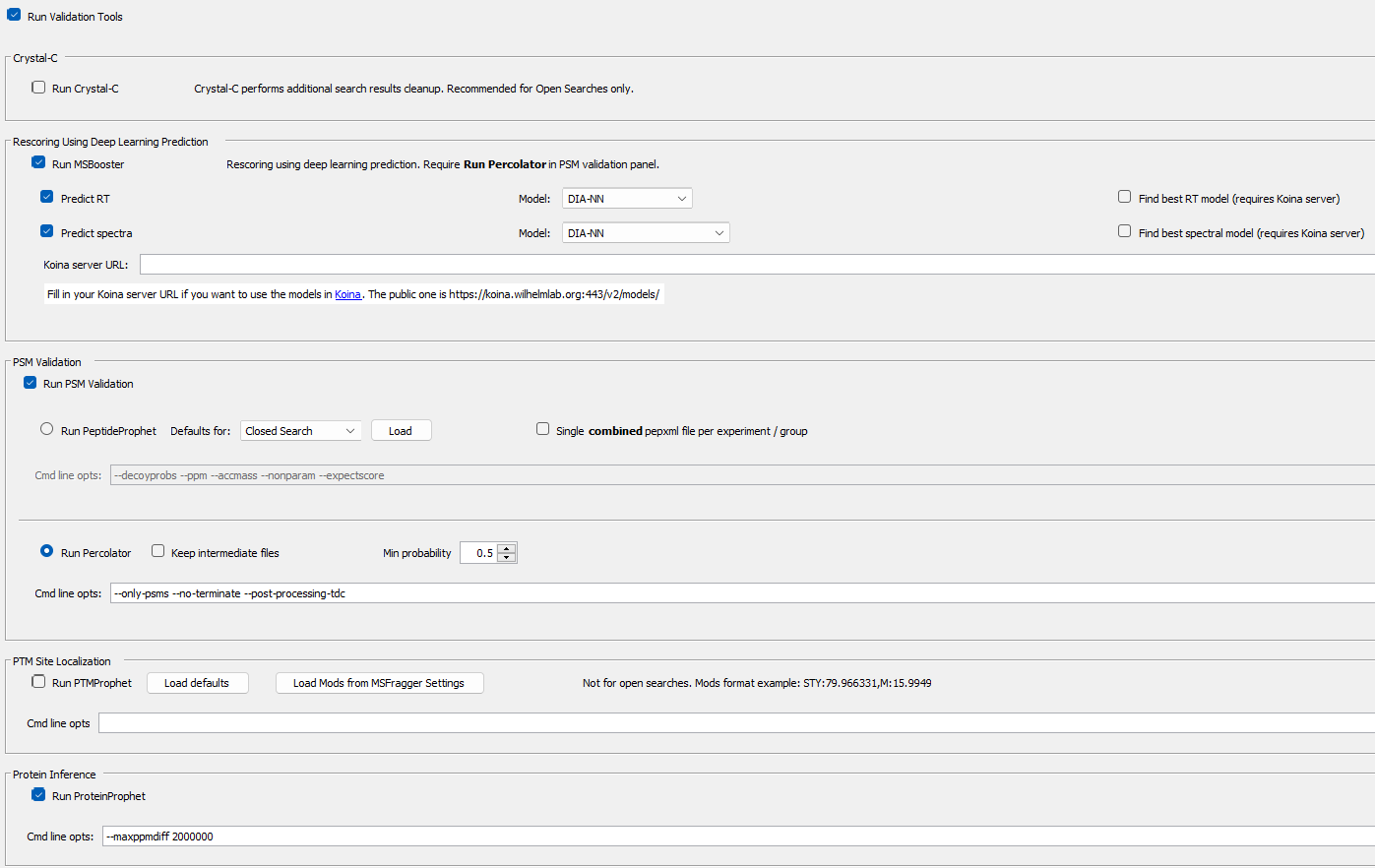 | ||
|
|
||
| 4. Set your output directory in the run tab to the Koina_MSBooster_tutorial folder | ||
|
|
||
| ### Running with the default DIA-NN model | ||
|
|
||
| The default prediction model for both MS2 spectra and RT in MSBooster is DIA-NN. If we run our workflow, we'll see | ||
| 2615 peptides passed 1% PSM and peptide FDR cutoffs. This number may vary slightly based on differences in software versions | ||
|
|
||
|
|
||
| ### Choosing a Koina model | ||
|
|
||
| We have our baseline performance at 2615 peptides with DIA-NN predictions. It is important to note that these peptides | ||
| were fragmented with CID instead of HCD. It is unclear whether DIA-NN was trained CID data or not. However, Prosit has | ||
| an explicit CID model. Let's swap in Prosit for DIA-NN. | ||
|
|
||
| Before using Koina, we must specify a Koina server URL. The publicly available one is | ||
| `https://koina.wilhelmlab.org:443/v2/models/`. Copy-paste this into the text box, if you are ok with sending out predictions | ||
| to an external server. Don't worry, the peptides submitted for prediction are not stored by the server! If you are working | ||
| with confidential data, consider [setting up your own Koina server](https://github.com/wilhelm-lab/koina). | ||
|
|
||
| Next, use the dropdown menus to choose the Prosit models. | ||
|
|
||
| 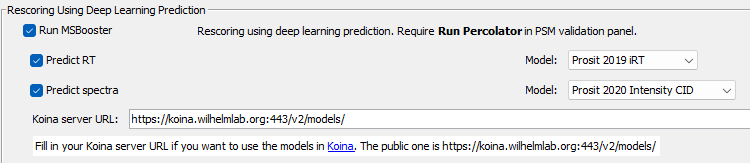 | ||
|
|
||
| Run this workflow now, and we get 2818 peptides. 200 more IDs! | ||
|
|
||
| ### Heuristic best model search | ||
|
|
||
| Let's say you don't know what models are best suited towards your data. There are some general guidelines you can follow | ||
| [here](https://www.biorxiv.org/content/10.1101/2024.06.01.596953v1) based on our tests, but as more models get added | ||
| to Koina, the number of models to choose from may become overwhelming. To address this, we have implemented a "best model | ||
| heuristic search" algorithm that tries to find the model best suited for you data. You can activate it by checking the | ||
| `Find best ___ model` checkboxes. Both DIA-NN and Koina models will be considered. | ||
|
|
||
|  | ||
|
|
||
| Let's look at the log to see what's going on: | ||
| ``` | ||
| ### RT model search | ||
| 2024-08-05 15:28:41 [INFO] - Searching for best RT model for your data | ||
| 2024-08-05 15:28:41 [INFO] - Searching the following models: | ||
| 2024-08-05 15:28:41 [INFO] - [DIA-NN, Deeplc_hela_hf, AlphaPept_rt_generic, Prosit_2019_irt, Prosit_2020_irt_TMT] | ||
| Iteration 1...2...3...4...5... | ||
| 2024-08-05 15:28:53 [INFO] - DIA-NN has root mean squared error of 3.5497 | ||
| Iteration 1...2...3...4...5... | ||
| 2024-08-05 15:28:56 [INFO] - Deeplc_hela_hf has root mean squared error of 3.3044 | ||
| Iteration 1...2...3...4...5... | ||
| 2024-08-05 15:28:57 [INFO] - AlphaPept_rt_generic has root mean squared error of 2.8290 | ||
| Iteration 1...2...3...4...5... | ||
| 2024-08-05 15:28:59 [INFO] - Prosit_2019_irt has root mean squared error of 3.7188 | ||
| Iteration 1...2...3...4...5... | ||
| 2024-08-05 15:29:00 [INFO] - Prosit_2020_irt_TMT has root mean squared error of 9.6557 | ||
| 2024-08-05 15:29:00 [INFO] - RT model chosen is AlphaPept_rt_generic | ||
| ``` | ||
|
|
||
| In the RT model search, MSBooster takes 1000 high-quality PSMs to perform RT calibration. While the root mean squared | ||
| error (RMSE) metric is shown, a "top consensus" method is used, described in more detail in our | ||
| [paper](https://www.biorxiv.org/content/10.1101/2024.06.01.596953v1). That is to say, the model with lowest RMSE may not | ||
| be the one chosen. Still, RMSE is an informative metric to know the dispersion of predicted RTs. Prosit TMT has the | ||
| largest RMSE, which is expected since we aren't working with TMT data. Ultimately, AlphaPeptDeep RT is chosen. | ||
|
|
||
| ``` | ||
| ### Spectral model search | ||
| 2024-08-05 15:29:00 [INFO] - Searching for best spectra model for your data | ||
| 2024-08-05 15:29:00 [INFO] - Searching the following models: | ||
| 2024-08-05 15:29:00 [INFO] - [DIA-NN, ms2pip_2021_HCD, AlphaPept_ms2_generic, Prosit_2019_intensity, Prosit_2023_intensity_timsTOF, Prosit_2020_intensity_CID, Prosit_2020_intensity_TMT, Prosit_2020_intensity_HCD] | ||
| 2024-08-05 15:29:11 [INFO] - Median similarity for DIA-NN is 0.8155 | ||
| 2024-08-05 15:29:13 [INFO] - Median similarity for ms2pip_2021_HCD is 0.7662 | ||
| 2024-08-05 15:29:13 [INFO] - Calibrating NCE | ||
| 2024-08-05 15:29:19 [INFO] - Best NCE for AlphaPept_ms2_generic after calibration is 20 | ||
| 2024-08-05 15:29:19 [INFO] - Median similarity for AlphaPept_ms2_generic is 0.8888 | ||
| 2024-08-05 15:29:19 [INFO] - Consider lowering minNCE below 20 | ||
| 2024-08-05 15:29:19 [INFO] - Calibrating NCE | ||
| 2024-08-05 15:29:29 [INFO] - Best NCE for Prosit_2019_intensity after calibration is 20 | ||
| 2024-08-05 15:29:29 [INFO] - Median similarity for Prosit_2019_intensity is 0.8777 | ||
| 2024-08-05 15:29:29 [INFO] - Consider lowering minNCE below 20 | ||
| 2024-08-05 15:29:29 [INFO] - Calibrating NCE | ||
| 2024-08-05 15:29:35 [INFO] - Best NCE for Prosit_2023_intensity_timsTOF after calibration is 20 | ||
| 2024-08-05 15:29:35 [INFO] - Median similarity for Prosit_2023_intensity_timsTOF is 0.8878 | ||
| 2024-08-05 15:29:35 [INFO] - Consider lowering minNCE below 20 | ||
| 2024-08-05 15:29:37 [INFO] - Median similarity for Prosit_2020_intensity_CID is 0.9598 | ||
| 2024-08-05 15:29:37 [INFO] - Calibrating NCE | ||
| 2024-08-05 15:29:44 [INFO] - Best NCE for Prosit_2020_intensity_TMT after calibration is 21 | ||
| 2024-08-05 15:29:44 [INFO] - Median similarity for Prosit_2020_intensity_TMT is 0.6300 | ||
| 2024-08-05 15:29:44 [INFO] - Calibrating NCE | ||
| 2024-08-05 15:29:50 [INFO] - Best NCE for Prosit_2020_intensity_HCD after calibration is 20 | ||
| 2024-08-05 15:29:50 [INFO] - Median similarity for Prosit_2020_intensity_HCD is 0.8918 | ||
| 2024-08-05 15:29:50 [INFO] - Consider lowering minNCE below 20 | ||
| 2024-08-05 15:29:50 [INFO] - Spectra model chosen is Prosit_2020_intensity_CID | ||
| ``` | ||
|
|
||
| Here we see details of the best MS2 model search. As expected, Prosit CID performs the best, since it is the only CID | ||
| model currently Koina. Additionally, we see that the optimal normalized collision energy (NCE) for prediction is below 20%. | ||
| This is something you can customize in the [MSBooster standalone version](https://github.com/Nesvilab/MSBooster/blob/master/Koina.md#command-line). | ||
|
|
||
| Ultimately, this further optimized model combination results in 2863 peptide IDs, ~50 peptides more than using Prosit's | ||
| RT model. Neat! | ||
|
|
||
| ### Visualizing predictions in FragPipe-PDV | ||
|
|
||
| Starting from v1.2.1., [FragPipe-PDV](https://github.com/Nesvilab/FragPipe-PDV) can be used to visualize mirror plots of | ||
| experimental vs predicted spectra for both DIA-NN and Koina models. The prediction file to load is determined by what | ||
| model you ran last (printed in msbooster_params.txt). Koina predictions are saved in the file ending in `koina.mgf`. | ||
| While DIA-NN does not predict y/b-1/2 ions, many of the Koina models do. Peep the y1 and y2 ions! | ||
|
|
||
| 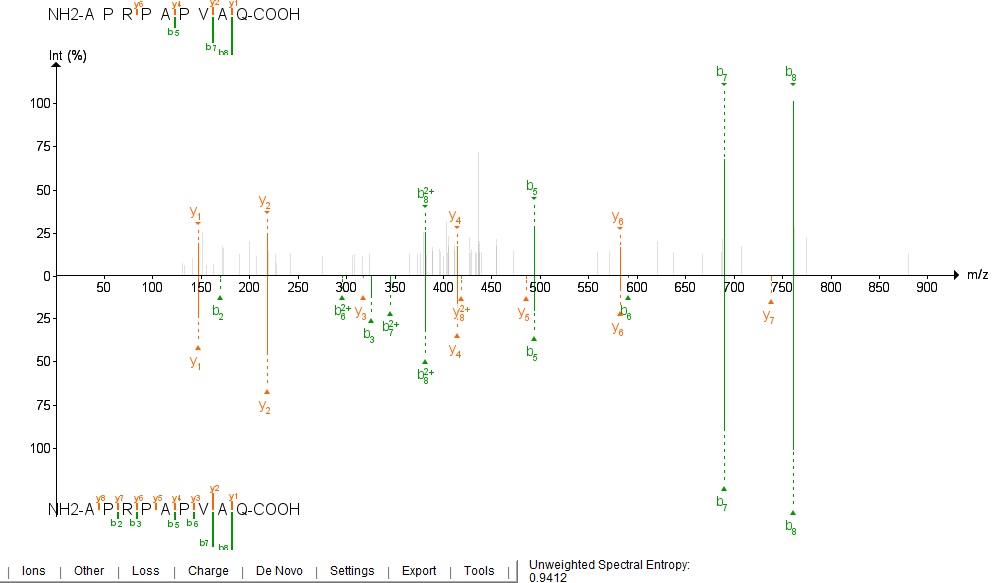 | ||
|
|
||
| ## Summary | ||
| MSBooster uses the Koina API to gain access to many more prediction models, leading to increased peptide IDs when there | ||
| is a better model than DIA-NN. Please refer to the [MSBooster documentation](https://github.com/Nesvilab/MSBooster/blob/master/Koina.md) | ||
| for more details, such as what QC figures are generated when Koina is run. | ||
|
|
||
| ## References | ||
| - <strong>MSBooster paper</strong>: Yang KL, Yu F, Teo GC, Li K, Demichev V, Ralser M, Nesvizhskii AI. <strong>MSBooster: | ||
| improving peptide identification rates using deep learning-based features</strong>. Nat Commun. 2023 Jul 27;14(1):4539. | ||
| doi: https://doi.org/10.1038%2Fs41467-023-40129-9. PMID: 37500632; PMCID: PMC10374903. | ||
| - <strong>Koina preprint</strong>: Lautenbacher L, Yang KL, Kockmann T, Panse C, Chambers M, Kahl E, Yu F, Gabriel W, | ||
| Bold D, Schmidt T, Li K, MacLean B, Nesvizhskii AI, Wilhelm M. <strong>Koina: Democratizing machine learning for proteomics | ||
| research</strong>. bioRxiv [Preprint]. 2024 Jun 3:2024.06.01.596953. doi: https://doi.org/10.1101%2F2024.06.01.596953. | ||
| PMID: 38895358; PMCID: PMC11185529. | ||
| - <strong>Data used in tutorial</strong>: Marcu A, Bichmann L, Kuchenbecker L, Kowalewski DJ, Freudenmann LK, Backert L, | ||
| Mühlenbruch L, Szolek A, Lübke M, Wagner P, Engler T, Matovina S, Wang J, Hauri-Hohl M, Martin R, Kapolou K, Walz JS, | ||
| Velz J, Moch H, Regli L, Silginer M, Weller M, Löffler MW, Erhard F, Schlosser A, Kohlbacher O, Stevanović S, Rammensee HG, | ||
| Neidert MC. <strong>HLA Ligand Atlas: a benign reference of HLA-presented peptides to improve T-cell-based cancer immunotherapy</strong>. | ||
| J Immunother Cancer. 2021 Apr;9(4):e002071. doi: https://doi.org/10.1136%2Fjitc-2020-002071. PMID: 33858848; PMCID: PMC8054196. | ||
|
|
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.