This project simulates the reaction kinetics of the reaction between an engineered leaf-branch compost cutinase and various polymers using the following differential equations
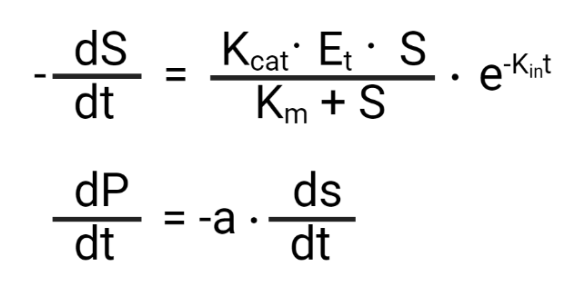
The dS/dt and dP/dt represent the substrate and product over time differential equations, respectively.

It then optimizes the actual data results to the model to find the most representative kinetic values.
-
Notifications
You must be signed in to change notification settings - Fork 0
TheGordon/Substrate-Enzyme-Simulation
Folders and files
| Name | Name | Last commit message | Last commit date | |
|---|---|---|---|---|
Repository files navigation
About
An archive of the substrate-enzyme simulation modeling for enzyme degradation research.
Resources
Stars
Watchers
Forks
Releases
No releases published
Packages 0
No packages published