The work is done at Vimslab in collaboration with DBI.
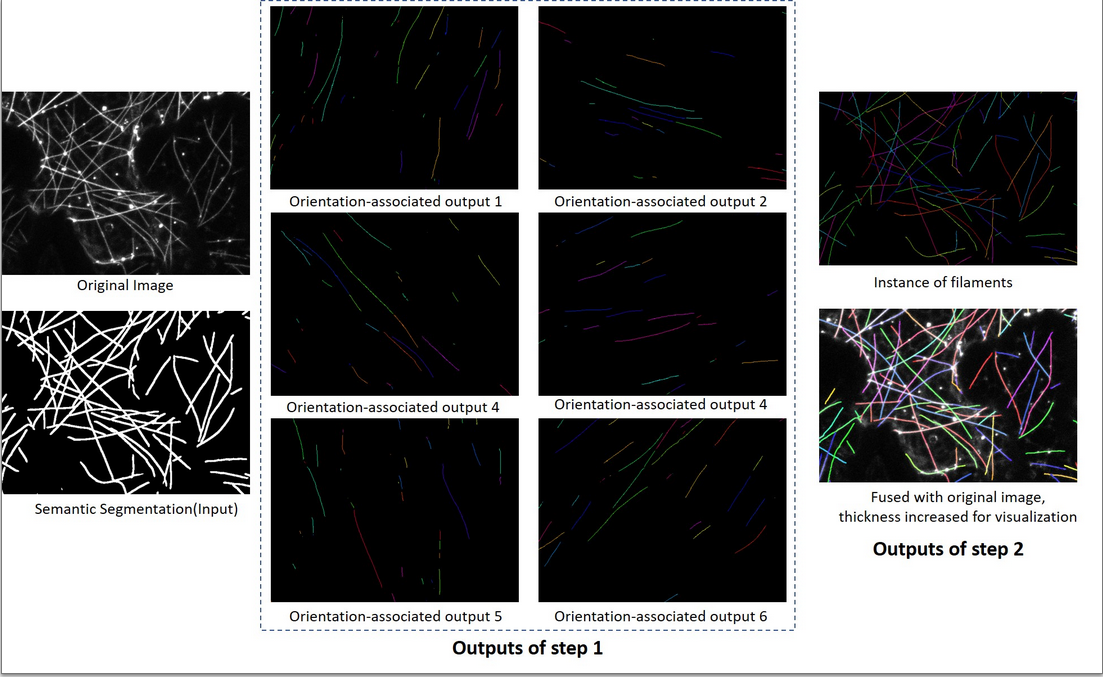
This project includes segmenation, seperation, reconnection three steps as stated in the paper. The proposed method aovid reconnecting filaments at intersection and thus acheives a better result than existing methods.
This repo is based on the following two papers:
- Liu, Y., Treible, W., Kolagunda, A., Nedo, A., Saponaro, P., Caplan, J. and Kambhamettu, C., Densely Connected Stacked U-Network for Filament Segmentation in Microscopy Images, ECCV Workshops, 2018. link
- Yi Liu, Abhishek Kolagunda, Wayne Treible, Alex Nedo, Jeffrey Caplan, Chandra Kambhamettu, Intersection To Overpass: Instance Segmentation on Filamentous Structures with An Orientation-Aware Neural Network and erminus Pairing Algorithm, CVPR Bioimaging Workshop, 2019. link
Anaconda enviroment file mtquant.yml
Matlab for reonnection step (Python version coming out soon)
We have collected three dataset and listed as follows
Binary Segmentation
- Microtubule dataset
- Actin dataset
Instance Label
- Microtubule instance dataset
Binary Segmentation
- Modify config.yaml file,
- Mode ts for timeSeries tif data Evaluation
- Mode train for training
- Mode predict for None timeSeries data evaluation
Separation by Orientation
- Modify config.yaml file,
- Mode train for training, Training should be conducted on customed synthetic dataset
- Mode predict for None timeSeries data evaluation
Reconnecting filaments
- run runMain_v2.sh
- If not using Slurm, check description in slurm_run_mat.py