code:https://github.com/RMeli/gnina-torch
manual: https://gnina-torch.readthedocs.io/en/latest/
1. create a index file (score.index) as following:
receptor_1.pdb ligand_1.sdf receptor_1.pdb ligand_2.sdf receptor_1.pdb ligand_3.sdf
2.rescore
python -m gninatorch.gnina \
score.index \
--batch_size 200 \
--cnn default
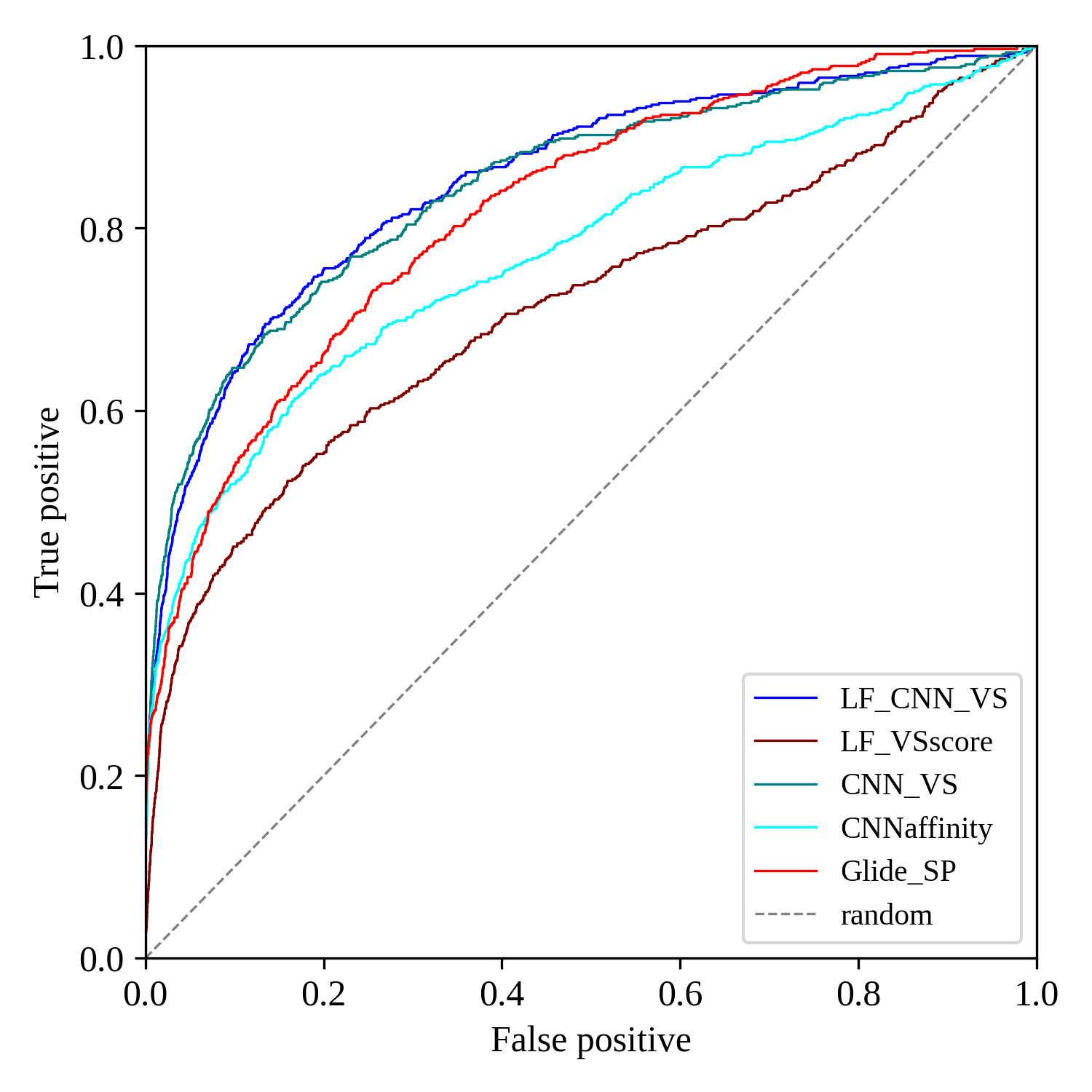
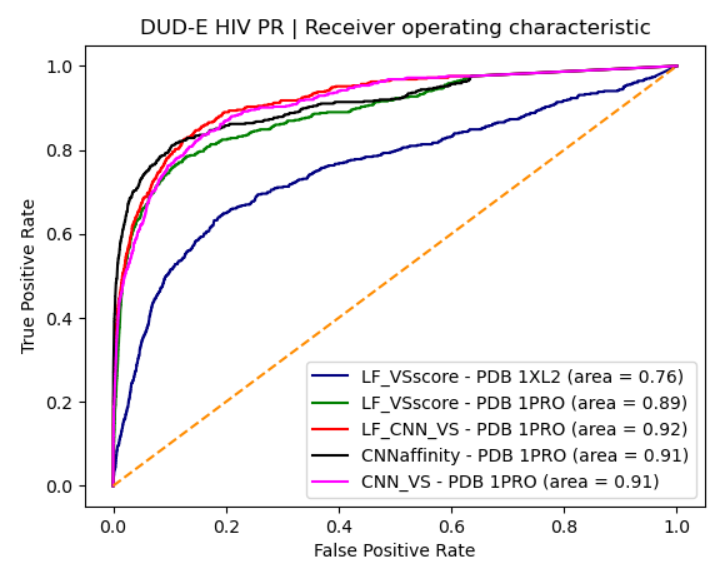
Blog: http://blog.molcalx.com.cn/2023/08/31/boosting-virtual-screening-enrichments-with-data-fusion.html
Blog: http://blog.molcalx.com.cn/2023/09/13/beware-of-the-benchmarking-test.html
- Sunseri, J.; Koes, D. R. Virtual Screening with Gnina 1.0. Molecules 2021, 26 (23), 7369. https://doi.org/10.3390/molecules26237369.
- https://github.com/RMeli/gnina-torch