Bayesian structural equation modelling with INLA.
Soon-ish features
- Model fit indices (PPP, xIC, RMSEA, etc.)
- Prior specification.
- Fixed values and/or parameter constraints.
- Specify different families for different observed variable.
- Standardised coefficients.
Long term plan
- “Non-iid” models, such as spatio-temporal models.
- Multilevel-ish kind of models (2-3 levels).
- Covariates.
- Multigroup analysis (in principle this is simple, but I have bigger plans for this).
- Missing data imputation.
You need a working installation of INLA. Following the official instructions given here, run this command in R:
install.packages(
"INLA",
repos = c(getOption("repos"),
INLA = "https://inla.r-inla-download.org/R/stable"),
dep = TRUE
)Then, you can install the development version of {INLAvaan} from
GitHub with:
# install.packages("pak")
pak::pak("haziqj/INLAvaan")A simple two-factor SEM with six observed, correlated Gaussian
variables. Let
For identifiability, we set
Graphically, we can plot the following path diagram.
# {lavaan} textual model
mod <- "
# Measurement model
eta1 =~ y1 + y2 + y3
eta2 =~ y4 + y5 + y6
# Factor regression
eta2 ~ eta1
# Covariances
y1 ~~ y4
y2 ~~ y5
y3 ~~ y6
"
# Data set
dplyr::glimpse(dat)
#> Rows: 10,000
#> Columns: 6
#> $ y1 <dbl> -1.21410027, 0.42781790, -1.23105577, 1.29706010, -0.14438098, -1.2…
#> $ y2 <dbl> -1.084814450, 0.372560944, -1.142733592, 1.906949830, -0.013115768,…
#> $ y3 <dbl> -1.7501130, 1.2281783, -0.9274464, 3.0957939, 0.7869644, -1.1736760…
#> $ y4 <dbl> 1.83828191, 0.92323072, 0.68456209, 1.88557378, -0.71608409, -0.519…
#> $ y5 <dbl> 1.32807356, 0.62454411, 1.26304139, 2.50170072, -0.82536783, -1.213…
#> $ y6 <dbl> 1.47070238, 1.36488688, 1.74260694, 2.89753599, -0.51635709, -0.748…To fit this model using {INLAvaan}, use the familiar {lavaan}
syntax. The i in isem stands for INLA (following the convention of
bsem for {blavaan}).
library(INLAvaan)
fit <- isem(model = mod, data = dat)
summary(fit)#> INLAvaan 0.1.0.9012 ended normally after 34 seconds
#>
#> Estimator BAYES
#> Optimization method INLA
#> Number of model parameters 16
#>
#> Number of observations 10000
#>
#> Statistic MargLogLik PPP
#> Value -52033.656 NA
#>
#> Parameter Estimates:
#>
#>
#> Latent Variables:
#> Estimate Post.SD pi.lower pi.upper Prior
#> eta1 =~
#> y1 1.000
#> y2 1.203 0.004 1.194 1.212 normal(0,10)
#> y3 1.496 0.005 1.485 1.505 normal(0,10)
#> eta2 =~
#> y4 1.000
#> y5 1.199 0.004 1.191 1.207 normal(0,10)
#> y6 1.503 0.005 1.493 1.512 normal(0,10)
#>
#> Regressions:
#> Estimate Post.SD pi.lower pi.upper Prior
#> eta2 ~
#> eta1 0.306 0.010 0.286 0.326 normal(0,10)
#>
#> Covariances:
#> Estimate Post.SD pi.lower pi.upper Prior
#> .y1 ~~
#> .y4 0.048 0.001 0.046 0.051 beta(1,1)
#> .y2 ~~
#> .y5 0.049 0.001 0.046 0.051 beta(1,1)
#> .y3 ~~
#> .y6 0.053 0.002 0.050 0.057 beta(1,1)
#>
#> Variances:
#> Estimate Post.SD pi.lower pi.upper Prior
#> .y1 0.100 0.002 0.096 0.103 gamma(1,.5)[sd]
#> .y2 0.100 0.002 0.096 0.104 gamma(1,.5)[sd]
#> .y3 0.103 0.003 0.097 0.108 gamma(1,.5)[sd]
#> .y4 0.098 0.002 0.094 0.101 gamma(1,.5)[sd]
#> .y5 0.098 0.002 0.094 0.103 gamma(1,.5)[sd]
#> .y6 0.105 0.003 0.099 0.111 gamma(1,.5)[sd]
#> eta1 1.009 0.015 0.980 1.040 gamma(1,.5)[sd]
#> .eta2 0.987 0.015 0.959 1.017 gamma(1,.5)[sd]
Compare model fit to {lavaan} and {blavaan} (MCMC sampling using
Stan on a single thread obtaining 1000 burnin and 2000 samples, as well
as variational Bayes):
#>
#> ── Compare timing (seconds) ──
#>
#> INLAvaan lavaan blavaan blavaan_vb
#> 34.968 0.028 127.584 92.434
A little experiment to see how sample size affects run time:
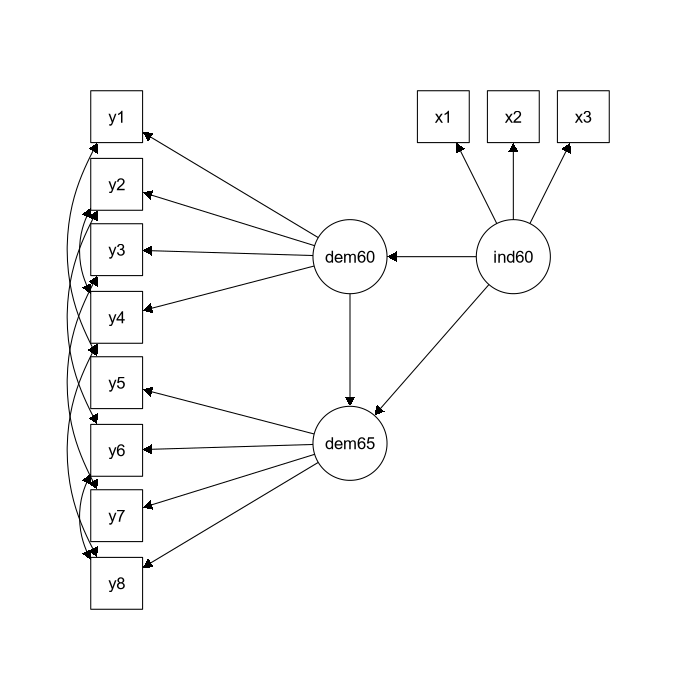
The quintessential example for SEM is this model from Bollen (1989) to fit a political democracy data set. Eleven observed variables are hypothesized to arise from three latent variables. This set includes data from 75 developing countries each assessed on four measures of democracy measured twice (1960 and 1965), and three measures of industrialization measured once (1960). The eleven observed variables are:
y1: Freedom of the press, 1960y2: Freedom of political opposition, 1960y3: Fairness of elections, 1960y4: Effectiveness of elected legislature, 1960y5: Freedom of the press, 1965y6: Freedom of political opposition, 1965y7: Fairness of elections, 1965y8: Effectiveness of elected legislature, 1965y9: GNP per capita, 1960y10: Energy consumption per capita, 1960y11: Percentage of labor force in industry, 1960
Variables y1-y4 and y5-y8 are typically used as indicators of the
latent trait of “political democracy” in 1960 and 1965 respectively,
whereas y9-y11 are used as indicators of industrialization (1960). It
is theorised that industrialisation influences political democracy, and
that political democracy in 1960 influences political democracy in 1965.
Since the items measure the same latent trait at two time points, there
is an assumption that the residuals of these items will be correlated
with each other. The model is depicted in the figure below.
The corresponding model in {lavaan} syntax is:
mod <- "
# latent variables
dem60 =~ y1 + y2 + y3 + y4
dem65 =~ y5 + y6 + y7 + y8
ind60 =~ x1 + x2 + x3
# latent regressions
dem60 ~ ind60
dem65 ~ ind60 + dem60
# residual covariances
y1 ~~ y5
y2 ~~ y4 + y6
y3 ~~ y7
y4 ~~ y8
y6 ~~ y8
"We will fit this model using {INLAvaan} and compare the results with
{blavaan}.
data("PoliticalDemocracy", package = "lavaan")
poldemfit <- isem(model = mod, data = PoliticalDemocracy)
summary(poldemfit)#> INLAvaan 0.1.0.9012 ended normally after 9 seconds
#>
#> Estimator BAYES
#> Optimization method INLA
#> Number of model parameters 31
#>
#> Number of observations 75
#>
#> Statistic MargLogLik PPP
#> Value -1607.444 NA
#>
#> Parameter Estimates:
#>
#>
#> Latent Variables:
#> Estimate Post.SD pi.lower pi.upper Prior
#> dem60 =~
#> y1 1.000
#> y2 1.256 0.184 0.894 1.620 normal(0,10)
#> y3 1.056 0.149 0.764 1.349 normal(0,10)
#> y4 1.262 0.150 0.967 1.558 normal(0,10)
#> dem65 =~
#> y5 1.000
#> y6 1.189 0.172 0.850 1.529 normal(0,10)
#> y7 1.281 0.162 0.963 1.599 normal(0,10)
#> y8 1.267 0.164 0.944 1.591 normal(0,10)
#> ind60 =~
#> x1 1.000
#> x2 2.186 0.138 1.923 2.465 normal(0,10)
#> x3 1.816 0.152 1.520 2.118 normal(0,10)
#>
#> Regressions:
#> Estimate Post.SD pi.lower pi.upper Prior
#> dem60 ~
#> ind60 1.478 0.398 0.691 2.259 normal(0,10)
#> dem65 ~
#> ind60 0.562 0.230 0.106 1.011 normal(0,10)
#> dem60 0.847 0.097 0.662 1.043 normal(0,10)
#>
#> Covariances:
#> Estimate Post.SD pi.lower pi.upper Prior
#> .y1 ~~
#> .y5 0.622 0.331 -0.042 1.234 beta(1,1)
#> .y2 ~~
#> .y4 1.372 0.604 0.306 2.743 beta(1,1)
#> .y6 2.183 0.664 1.025 3.479 beta(1,1)
#> .y3 ~~
#> .y7 0.770 0.560 -0.447 1.879 beta(1,1)
#> .y4 ~~
#> .y8 0.337 0.461 -0.594 1.259 beta(1,1)
#> .y6 ~~
#> .y8 1.427 0.488 0.502 2.339 beta(1,1)
#>
#> Variances:
#> Estimate Post.SD pi.lower pi.upper Prior
#> .y1 2.026 0.486 1.151 3.186 gamma(1,.5)[sd]
#> .y2 7.436 1.375 5.257 10.529 gamma(1,.5)[sd]
#> .y3 5.242 0.950 3.629 7.278 gamma(1,.5)[sd]
#> .y4 3.368 0.788 2.065 5.190 gamma(1,.5)[sd]
#> .y5 2.435 0.525 1.602 3.590 gamma(1,.5)[sd]
#> .y6 5.128 0.951 3.671 7.163 gamma(1,.5)[sd]
#> .y7 3.733 0.797 2.441 5.373 gamma(1,.5)[sd]
#> .y8 3.464 0.791 2.149 5.219 gamma(1,.5)[sd]
#> .x1 0.087 0.021 0.055 0.130 gamma(1,.5)[sd]
#> .x2 0.112 0.058 0.028 0.233 gamma(1,.5)[sd]
#> .x3 0.481 0.091 0.329 0.690 gamma(1,.5)[sd]
#> .dem60 4.118 0.982 2.518 6.356 gamma(1,.5)[sd]
#> .dem65 0.104 0.180 -0.002 0.560 gamma(1,.5)[sd]
#> ind60 0.462 0.090 0.310 0.662 gamma(1,.5)[sd]
#>
#> ── Compare timing (seconds) ──
#>
#> INLAvaan blavaan
#> 9.004 19.669
sessioninfo::session_info(info = "all")
#> ─ Session info ───────────────────────────────────────────────────────────────
#> setting value
#> version R version 4.4.0 (2024-04-24)
#> os macOS Sonoma 14.5
#> system aarch64, darwin20
#> ui X11
#> language (EN)
#> collate en_US.UTF-8
#> ctype en_US.UTF-8
#> tz Asia/Brunei
#> date 2024-06-21
#> pandoc 3.2 @ /opt/homebrew/bin/ (via rmarkdown)
#>
#> ─ Packages ───────────────────────────────────────────────────────────────────
#> package * version date (UTC) lib source
#> abind 1.4-5 2016-07-21 [1] CRAN (R 4.4.0)
#> arm 1.14-4 2024-04-01 [1] CRAN (R 4.4.0)
#> backports 1.5.0 2024-05-23 [1] CRAN (R 4.4.0)
#> base64enc 0.1-3 2015-07-28 [1] CRAN (R 4.4.0)
#> bayesplot 1.11.1 2024-02-15 [1] CRAN (R 4.4.0)
#> blavaan * 0.5-5 2024-06-09 [1] CRAN (R 4.4.0)
#> boot 1.3-30 2024-02-26 [1] CRAN (R 4.4.0)
#> carData 3.0-5 2022-01-06 [1] CRAN (R 4.4.0)
#> checkmate 2.3.1 2023-12-04 [1] CRAN (R 4.4.0)
#> class 7.3-22 2023-05-03 [1] CRAN (R 4.4.0)
#> classInt 0.4-10 2023-09-05 [1] CRAN (R 4.4.0)
#> cli 3.6.2 2023-12-11 [1] CRAN (R 4.4.0)
#> clue 0.3-65 2023-09-23 [1] CRAN (R 4.4.0)
#> cluster 2.1.6 2023-12-01 [1] CRAN (R 4.4.0)
#> coda 0.19-4.1 2024-01-31 [1] CRAN (R 4.4.0)
#> codetools 0.2-20 2024-03-31 [1] CRAN (R 4.4.0)
#> colorspace 2.1-0 2023-01-23 [1] CRAN (R 4.4.0)
#> CompQuadForm 1.4.3 2017-04-12 [1] CRAN (R 4.4.0)
#> corpcor 1.6.10 2021-09-16 [1] CRAN (R 4.4.0)
#> curl 5.2.1 2024-03-01 [1] CRAN (R 4.4.0)
#> data.table 1.15.4 2024-03-30 [1] CRAN (R 4.4.0)
#> DBI 1.2.3 2024-06-02 [1] CRAN (R 4.4.0)
#> digest 0.6.35 2024-03-11 [1] CRAN (R 4.4.0)
#> dplyr * 1.1.4 2023-11-17 [1] CRAN (R 4.4.0)
#> e1071 1.7-14 2023-12-06 [1] CRAN (R 4.4.0)
#> evaluate 0.24.0 2024-06-10 [1] CRAN (R 4.4.0)
#> fansi 1.0.6 2023-12-08 [1] CRAN (R 4.4.0)
#> farver 2.1.2 2024-05-13 [1] CRAN (R 4.4.0)
#> fastmap 1.2.0 2024-05-15 [1] CRAN (R 4.4.0)
#> fBasics 4032.96 2023-11-03 [1] CRAN (R 4.4.0)
#> fdrtool 1.2.17 2021-11-13 [1] CRAN (R 4.4.0)
#> fmesher 0.1.6 2024-06-14 [1] CRAN (R 4.4.0)
#> forcats * 1.0.0 2023-01-29 [1] CRAN (R 4.4.0)
#> foreign 0.8-86 2023-11-28 [1] CRAN (R 4.4.0)
#> Formula 1.2-5 2023-02-24 [1] CRAN (R 4.4.0)
#> future 1.33.2 2024-03-26 [1] CRAN (R 4.4.0)
#> future.apply 1.11.2 2024-03-28 [1] CRAN (R 4.4.0)
#> generics 0.1.3 2022-07-05 [1] CRAN (R 4.4.0)
#> ggplot2 * 3.5.1 2024-04-23 [1] CRAN (R 4.4.0)
#> glasso 1.11 2019-10-01 [1] CRAN (R 4.4.0)
#> globals 0.16.3 2024-03-08 [1] CRAN (R 4.4.0)
#> glue 1.7.0 2024-01-09 [1] CRAN (R 4.4.0)
#> gridExtra 2.3 2017-09-09 [1] CRAN (R 4.4.0)
#> gt 0.10.1 2024-01-17 [1] CRAN (R 4.4.0)
#> gtable 0.3.5 2024-04-22 [1] CRAN (R 4.4.0)
#> gtools 3.9.5 2023-11-20 [1] CRAN (R 4.4.0)
#> highr 0.11 2024-05-26 [1] CRAN (R 4.4.0)
#> Hmisc 5.1-3 2024-05-28 [1] CRAN (R 4.4.0)
#> hms 1.1.3 2023-03-21 [1] CRAN (R 4.4.0)
#> htmlTable 2.4.2 2023-10-29 [1] CRAN (R 4.4.0)
#> htmltools 0.5.8.1 2024-04-04 [1] CRAN (R 4.4.0)
#> htmlwidgets 1.6.4 2023-12-06 [1] CRAN (R 4.4.0)
#> igraph 2.0.3 2024-03-13 [1] CRAN (R 4.4.0)
#> INLA 24.06.04 2024-06-03 [1] local
#> INLAvaan * 0.1.0.9013 2024-05-31 [1] local
#> inline 0.3.19 2021-05-31 [1] CRAN (R 4.4.0)
#> jpeg 0.1-10 2022-11-29 [1] CRAN (R 4.4.0)
#> jsonlite 1.8.8 2023-12-04 [1] CRAN (R 4.4.0)
#> KernSmooth 2.23-24 2024-05-17 [1] CRAN (R 4.4.0)
#> knitr 1.47 2024-05-29 [1] CRAN (R 4.4.0)
#> kutils 1.73 2023-09-17 [1] CRAN (R 4.4.0)
#> labeling 0.4.3 2023-08-29 [1] CRAN (R 4.4.0)
#> lattice 0.22-6 2024-03-20 [1] CRAN (R 4.4.0)
#> lavaan * 0.6-18 2024-06-07 [1] CRAN (R 4.4.0)
#> lifecycle 1.0.4 2023-11-07 [1] CRAN (R 4.4.0)
#> lisrelToR 0.3 2024-02-07 [1] CRAN (R 4.4.0)
#> listenv 0.9.1 2024-01-29 [1] CRAN (R 4.4.0)
#> lme4 1.1-35.4 2024-06-19 [1] CRAN (R 4.4.0)
#> loo 2.7.0 2024-02-24 [1] CRAN (R 4.4.0)
#> lubridate * 1.9.3 2023-09-27 [1] CRAN (R 4.4.0)
#> magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.4.0)
#> MASS 7.3-61 2024-06-13 [1] CRAN (R 4.4.0)
#> Matrix 1.7-0 2024-03-22 [1] CRAN (R 4.4.0)
#> matrixStats 1.3.0 2024-04-11 [1] CRAN (R 4.4.0)
#> mgcv 1.9-1 2023-12-21 [1] CRAN (R 4.4.0)
#> mi 1.1 2022-06-06 [1] CRAN (R 4.4.0)
#> minqa 1.2.7 2024-05-20 [1] CRAN (R 4.4.0)
#> mnormt 2.1.1 2022-09-26 [1] CRAN (R 4.4.0)
#> modeest 2.4.0 2019-11-18 [1] CRAN (R 4.4.0)
#> munsell 0.5.1 2024-04-01 [1] CRAN (R 4.4.0)
#> mvtnorm 1.2-5 2024-05-21 [1] CRAN (R 4.4.0)
#> nlme 3.1-165 2024-06-06 [1] CRAN (R 4.4.0)
#> nloptr 2.1.0 2024-06-19 [1] CRAN (R 4.4.0)
#> nnet 7.3-19 2023-05-03 [1] CRAN (R 4.4.0)
#> nonnest2 0.5-7 2024-05-06 [1] CRAN (R 4.4.0)
#> OpenMx 2.21.11 2023-11-28 [1] CRAN (R 4.4.0)
#> openxlsx 4.2.5.2 2023-02-06 [1] CRAN (R 4.4.0)
#> parallelly 1.37.1 2024-02-29 [1] CRAN (R 4.4.0)
#> pbapply 1.7-2 2023-06-27 [1] CRAN (R 4.4.0)
#> pbivnorm 0.6.0 2015-01-23 [1] CRAN (R 4.4.0)
#> pillar 1.9.0 2023-03-22 [1] CRAN (R 4.4.0)
#> pkgbuild 1.4.4 2024-03-17 [1] CRAN (R 4.4.0)
#> pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.4.0)
#> plyr 1.8.9 2023-10-02 [1] CRAN (R 4.4.0)
#> png 0.1-8 2022-11-29 [1] CRAN (R 4.4.0)
#> proxy 0.4-27 2022-06-09 [1] CRAN (R 4.4.0)
#> psych 2.4.3 2024-03-18 [1] CRAN (R 4.4.0)
#> purrr * 1.0.2 2023-08-10 [1] CRAN (R 4.4.0)
#> qgraph 1.9.8 2023-11-03 [1] CRAN (R 4.4.0)
#> quadprog 1.5-8 2019-11-20 [1] CRAN (R 4.4.0)
#> QuickJSR 1.2.2 2024-06-07 [1] CRAN (R 4.4.0)
#> R6 2.5.1 2021-08-19 [1] CRAN (R 4.4.0)
#> Rcpp * 1.0.12 2024-01-09 [1] CRAN (R 4.4.0)
#> RcppParallel 5.1.7 2023-02-27 [1] CRAN (R 4.4.0)
#> readr * 2.1.5 2024-01-10 [1] CRAN (R 4.4.0)
#> reshape2 1.4.4 2020-04-09 [1] CRAN (R 4.4.0)
#> rlang 1.1.4 2024-06-04 [1] CRAN (R 4.4.0)
#> rmarkdown 2.27 2024-05-17 [1] CRAN (R 4.4.0)
#> rmutil 1.1.10 2022-10-27 [1] CRAN (R 4.4.0)
#> rockchalk 1.8.157 2022-08-06 [1] CRAN (R 4.4.0)
#> rpart 4.1.23 2023-12-05 [1] CRAN (R 4.4.0)
#> rstan 2.32.6 2024-03-05 [1] CRAN (R 4.4.0)
#> rstantools 2.4.0 2024-01-31 [1] CRAN (R 4.4.0)
#> rstudioapi 0.16.0 2024-03-24 [1] CRAN (R 4.4.0)
#> sandwich 3.1-0 2023-12-11 [1] CRAN (R 4.4.0)
#> scales 1.3.0 2023-11-28 [1] CRAN (R 4.4.0)
#> sem 3.1-15 2022-04-10 [1] CRAN (R 4.4.0)
#> semPlot * 1.1.6 2022-08-10 [1] CRAN (R 4.4.0)
#> semptools * 0.2.10 2023-10-15 [1] CRAN (R 4.4.0)
#> sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.4.0)
#> sf 1.0-16 2024-03-24 [1] CRAN (R 4.4.0)
#> sp 2.1-4 2024-04-30 [1] CRAN (R 4.4.0)
#> spatial 7.3-17 2023-07-20 [1] CRAN (R 4.4.0)
#> stable 1.1.6 2022-03-02 [1] CRAN (R 4.4.0)
#> stabledist 0.7-1 2016-09-12 [1] CRAN (R 4.4.0)
#> StanHeaders 2.32.9 2024-05-29 [1] CRAN (R 4.4.0)
#> statip 0.2.3 2019-11-17 [1] CRAN (R 4.4.0)
#> stringi 1.8.4 2024-05-06 [1] CRAN (R 4.4.0)
#> stringr * 1.5.1 2023-11-14 [1] CRAN (R 4.4.0)
#> tibble * 3.2.1 2023-03-20 [1] CRAN (R 4.4.0)
#> tidyr * 1.3.1 2024-01-24 [1] CRAN (R 4.4.0)
#> tidyselect 1.2.1 2024-03-11 [1] CRAN (R 4.4.0)
#> tidyverse * 2.0.0 2023-02-22 [1] CRAN (R 4.4.0)
#> timechange 0.3.0 2024-01-18 [1] CRAN (R 4.4.0)
#> timeDate 4032.109 2023-12-14 [1] CRAN (R 4.4.0)
#> timeSeries 4032.109 2024-01-14 [1] CRAN (R 4.4.0)
#> tmvnsim 1.0-2 2016-12-15 [1] CRAN (R 4.4.0)
#> tzdb 0.4.0 2023-05-12 [1] CRAN (R 4.4.0)
#> units 0.8-5 2023-11-28 [1] CRAN (R 4.4.0)
#> utf8 1.2.4 2023-10-22 [1] CRAN (R 4.4.0)
#> V8 4.4.2 2024-02-15 [1] CRAN (R 4.4.0)
#> vctrs 0.6.5 2023-12-01 [1] CRAN (R 4.4.0)
#> withr 3.0.0 2024-01-16 [1] CRAN (R 4.4.0)
#> xfun 0.45 2024-06-16 [1] CRAN (R 4.4.0)
#> XML 3.99-0.16.1 2024-01-22 [1] CRAN (R 4.4.0)
#> xml2 1.3.6 2023-12-04 [1] CRAN (R 4.4.0)
#> xtable 1.8-4 2019-04-21 [1] CRAN (R 4.4.0)
#> yaml 2.3.8 2023-12-11 [1] CRAN (R 4.4.0)
#> zip 2.3.1 2024-01-27 [1] CRAN (R 4.4.0)
#> zoo 1.8-12 2023-04-13 [1] CRAN (R 4.4.0)
#>
#> [1] /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/library
#>
#> ─ External software ──────────────────────────────────────────────────────────
#> setting value
#> cairo 1.17.6
#> cairoFT
#> pango 1.50.14
#> png 1.6.40
#> jpeg 9.5
#> tiff LIBTIFF, Version 4.5.0
#> tcl 8.6.13
#> curl 8.6.0
#> zlib 1.2.12
#> bzlib 1.0.8, 13-Jul-2019
#> xz 5.4.4
#> deflate
#> PCRE 10.42 2022-12-11
#> ICU 74.1
#> TRE TRE 0.8.0 R_fixes (BSD)
#> iconv Apple or GNU libiconv 1.11
#> readline 5.2
#> BLAS /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/lib/libRblas.0.dylib
#> lapack /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/lib/libRlapack.dylib
#> lapack_version 3.12.0
#>
#> ─ Python configuration ───────────────────────────────────────────────────────
#> Python is not available
#>
#> ──────────────────────────────────────────────────────────────────────────────