Learning SliDL, a Python library of pre- and post-processing tools for applying deep learning to whole-slide images
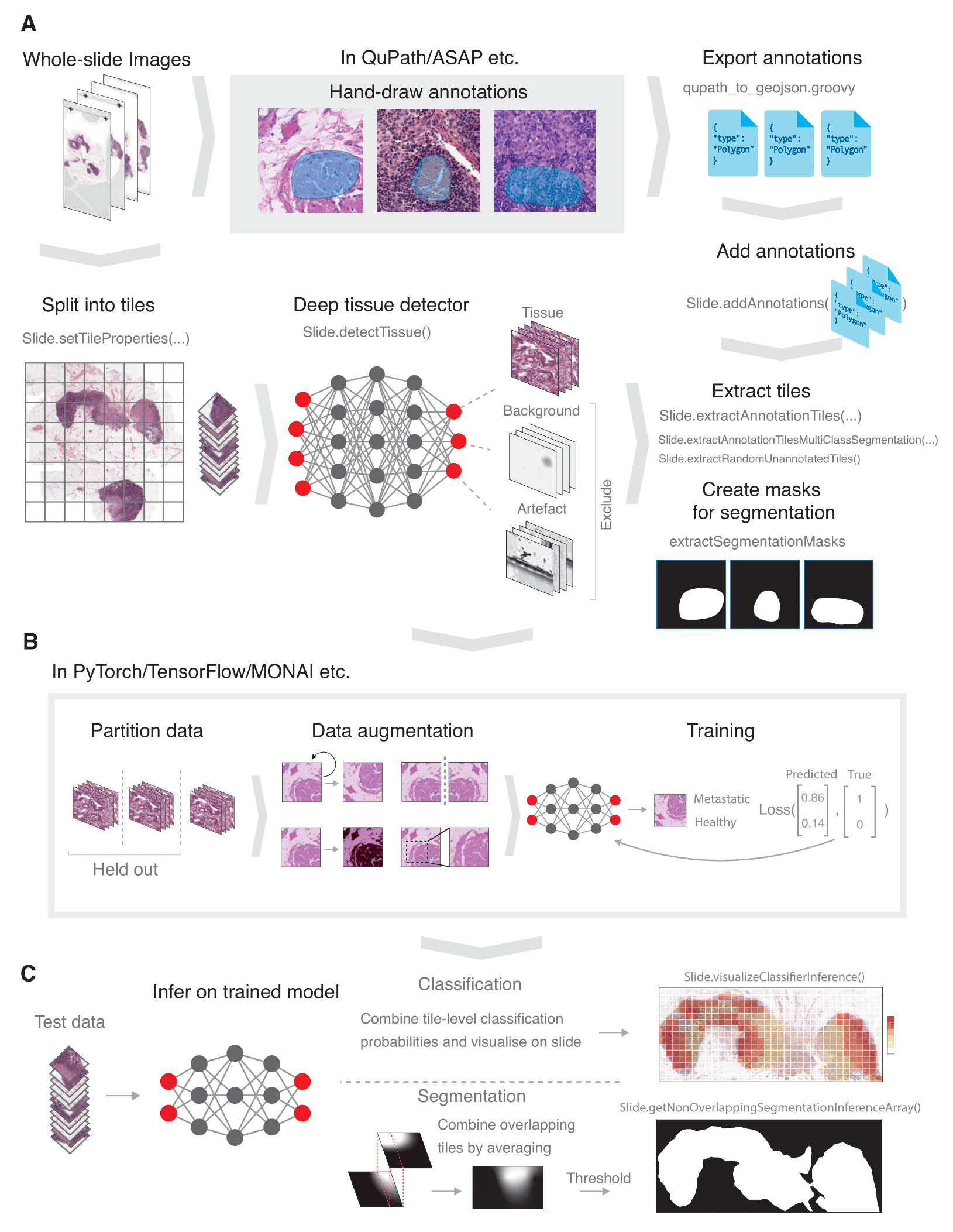
SliDL is a Python library for performing deep learning image analysis on whole-slide images (WSIs), including deep tissue, artefact, and background filtering, tile extraction, model inference, model evaluation and more. This repository serves to teach users how to apply SliDL on both a classification and a segmentation example problem from start to finish using best practices.
SliDL can also be installed via the Python Package Index (PyPI):
pip install slidl
First clone this repository:
git clone https://github.com/markowetzlab/slidl-tutorial
The tutorial uses an example subset lymph node WSIs from the CAMELYON16 challenge. Some of these WSIs contain breast cancer metastases and the goal of the tutorial is to use SliDL to train deep learning models to identify metastasis-containing slides and slide regions, and then to evaluate the performance of those models.
Create a directory called wsi_data where there is at least 38 GB of disk space. Download the following 18 WSIs from the CAMELYON16 dataset into wsi_data:
normal/normal_001.tifnormal/normal_010.tifnormal/normal_028.tifnormal/normal_037.tifnormal/normal_055.tifnormal/normal_074.tifnormal/normal_111.tifnormal/normal_141.tifnormal/normal_160.tiftumor/tumor_009.tiftumor/tumor_011.tiftumor/tumor_036.tiftumor/tumor_039.tiftumor/tumor_044.tiftumor/tumor_046.tiftumor/tumor_058.tiftumor/tumor_076.tiftumor/tumor_085.tif
Install Jupyter notebook into slidl-env:
conda install -c conda-forge notebook
Now that the requisite software and data have been downloaded, you are ready to begin the tutorial, which is contained in the Jupyter notebook slidl-tutorial.ipynb in this repository. Start notebook and then navigate to that document in the interface:
jupyter notebook
Once up and running, slidl-tutorial.ipynb contains instructions for running the tutorial. For instructions on running Jupyter notebooks, see the Jupyter documentation.
The results of a completed tutorial run can be found here.
The implementation of the U-Net segmentation architecture contained in this repository and some related segmentation code comes from milesial's open source project.
The complete documentation for SliDL including its API reference can be found here.
Note that this is prerelease software. Please use accordingly.