MGFIT (IDL version) - IDL/GDL Library for Least-Squares Minimization Genetic Algorithm Fitting
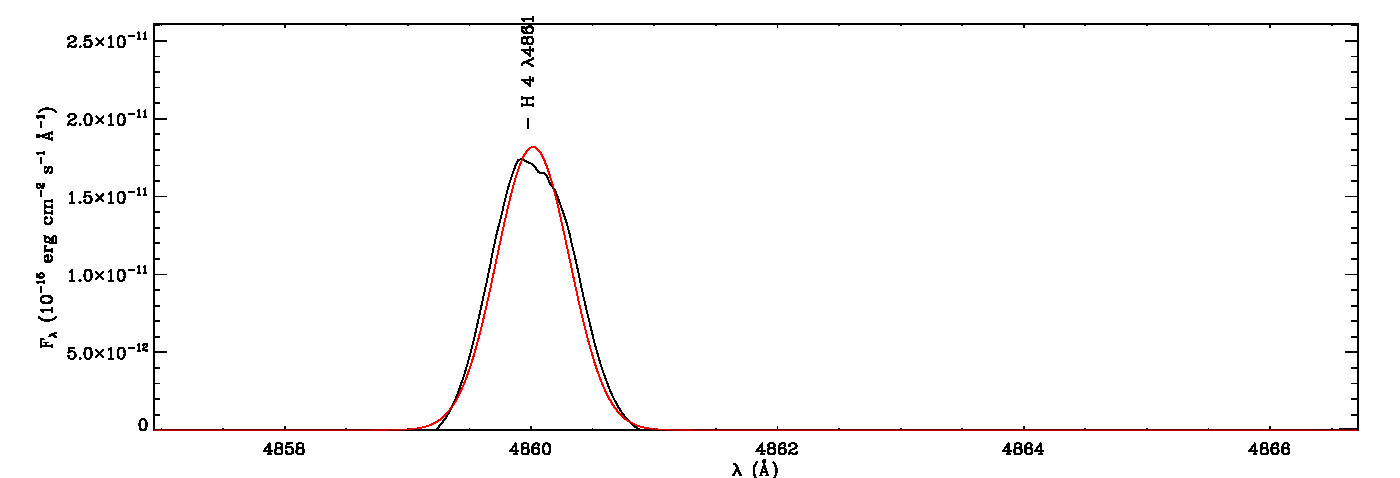
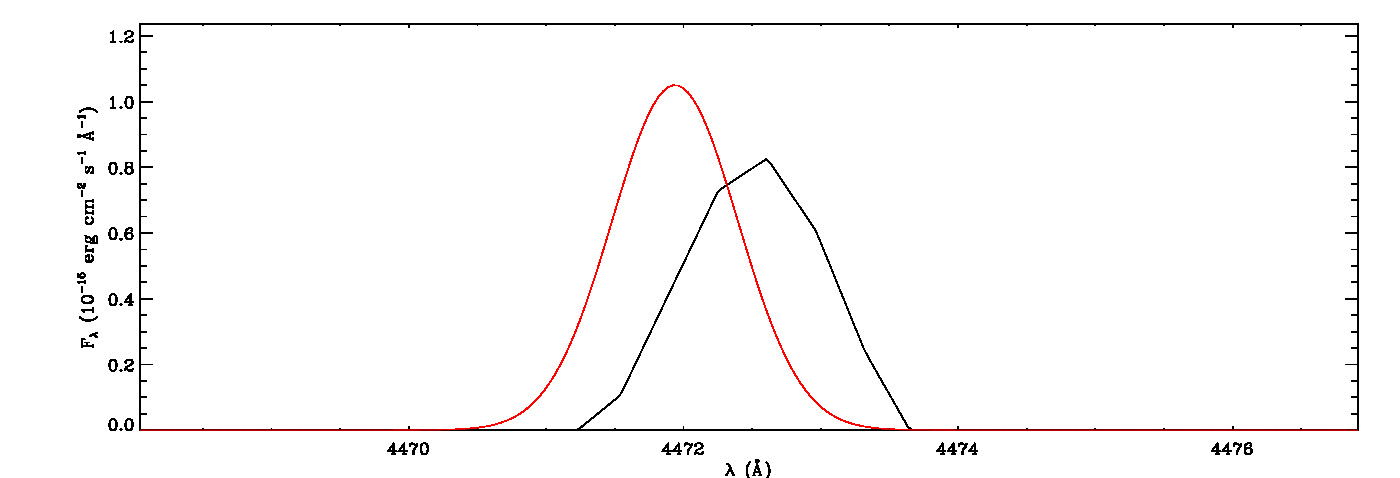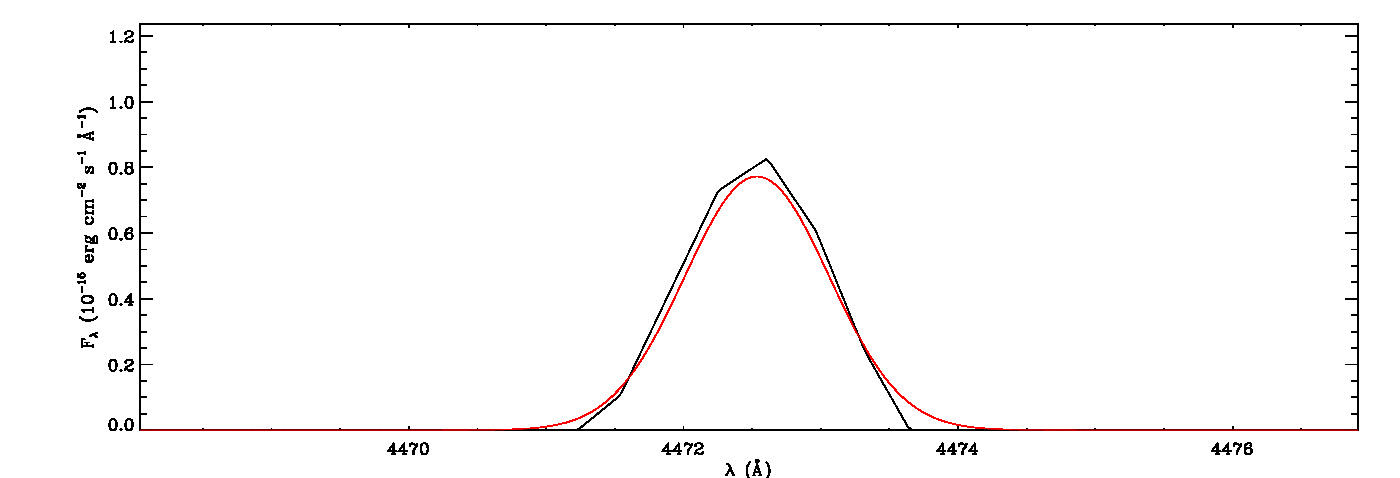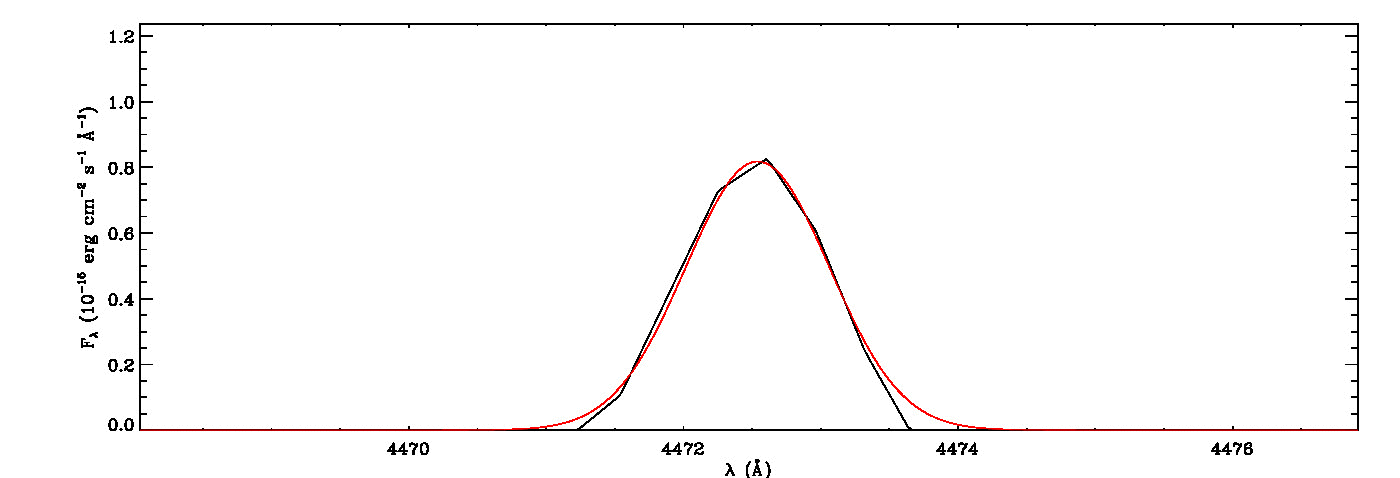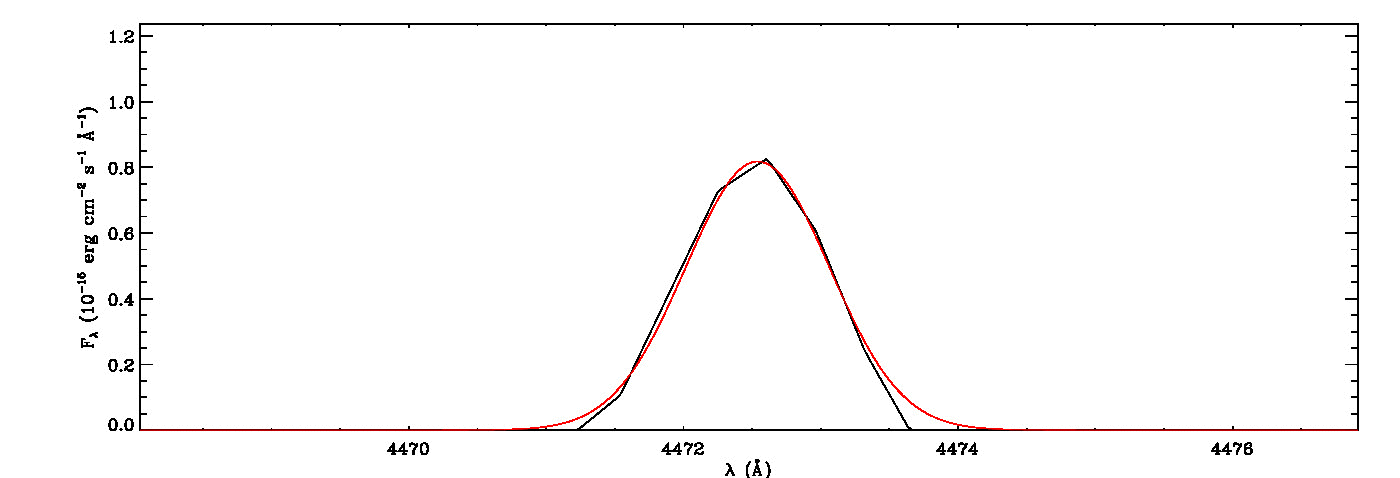
The MGFIT is an Interactive Data Language (IDL)/GNU Data Language (GDL) Library developed to fit multiple Gaussian functions to a list of emission (or absorption) lines using a least-squares minimization technique and a random walk method in three-dimensional locations of the specified lines, namely line peak, line width, and wavelength shift. It uses the MPFIT IDL Library (MINPACK-1 Least Squares Fitting; Markwardt 2009), which performs Levenberg-Marquardt least-squares minimization, to estimate the seed values required for initializing the three-dimensional coordination of each line in the first iteration. It then uses a random walk method optimized using a genetic algorithm originally evolved from the early version of the Fortran program ALFA (Automated Line Fitting Algorithm; Wesson 2016) to determine the best fitting values of the specified lines. The continuum curve is determined and subtracted before the line identification and flux measurements. It quantifies the white noise of the spectrum, which is then utilized to estimate uncertainties of fitted lines using the signal-dependent noise model of least-squares Gaussian fitting (Lenz & Ayres 1992) built on the work of Landman, Roussel-Dupre, and Tanigawa (1982).
This package requires the following packages:
To get this package with all the dependent packages, you can simply use
gitcommand as follows:git clone --recursive https://github.com/mgfit/MGFIT-idl
- To install the MGFIT IDL library in the Interactive Data Language (IDL), you need to add the path of this package directory to your IDL path. For more information about the path management in IDL, read the tips for customizing IDL program path provided by Harris Geospatial Solutions or the IDL library installation note by David Fanning in the Coyote IDL Library.
- This package requires IDL version 7.1 or later.
You can install the GNU Data Language (GDL) if you do not have it on your machine:
Linux (Fedora):
sudo dnf install gdl
Linux (Ubuntu):
sudo apt-get install gnudatalanguage
OS X (brew):
brew tap brewsci/science brew install gnudatalanguage
OS X (macports):
sudo port selfupdate sudo port upgrade libtool sudo port install gnudatalanguage
Windows: You can use the GNU Data Language for Win32 (Unofficial Version) or you can compile the GitHub source using Visual Studio 2015 as shown in appveyor.yml.
To install the MGFIT library in GDL, you need to add the path of this package directory to your
.gdl_startupfile in your home directory:!PATH=!PATH + ':/home/MGFIT-idl/pro/' !PATH=!PATH + ':/home/MGFIT-idl/externals/astron/pro/' !PATH=!PATH + ':/home/MGFIT-idl/externals/coyote/pro/' !PATH=!PATH + ':/home/MGFIT-idl/externals/coyote/public/' !PATH=!PATH + ':/home/MGFIT-idl/externals/mpfit/' !PATH=!PATH + ':/home/MGFIT-idl/externals/textoidl/'
You may also need to set
GDL_STARTUPif you have not done in.bashrc(bash):export GDL_STARTUP=~/.gdl_startup
or in
.tcshrc(cshrc):setenv GDL_STARTUP ~/.gdl_startup
This package requires GDL version 0.9.9 or later.
The Documentation of the IDL functions provides in detail in the API Documentation (mgfit.github.io/MGFIT-idl/doc). This IDL library fit multiple Gaussian functions to a list of emission lines in the given input spectrum.
You need to load the line list database:
base_dir = file_dirname(file_dirname((routine_info('$MAIN$', /source)).path))
data_dir = ['data']
fits_file = filepath('linedata.fits', root_dir=base_dir, subdir=data_dir )
strongline_data=read_stronglines(fits_file)
deepline_data=read_deeplines(fits_file)also load your spectrum arrays: wavelength array, and flux array (see examples):
input_dir = ['examples','inputs']
input_file = filepath('spectrum.txt', root_dir=base_dir, subdir=input_dir )
mgfit_read_ascii, input_file, wavel, fluxand define the output paths:
output_dir = ['examples','outputs']
image_dir = ['examples','images']
image_output_path = filepath('', root_dir=base_dir, subdir=image_dir )
output_path = filepath('', root_dir=base_dir, subdir=output_dir )You need to specify the genetic algorithm settings:
popsize=30.
pressure=0.3
generations=500.and use the appropriate fitting settings such as the wavelength interval, the redshift, and the spectral FWHM:
interval_wavelength=500
redshift_initial = 1.0
redshift_tolerance=0.001
fwhm_initial=1.0
fwhm_tolerance=1.4
fwhm_min=0.1
fwhm_max=1.8Now you run the MGFIT main function as follows:
emissionlines = mgfit_detect_lines(wavel, flux, deepline_data, strongline_data, $
popsize=popsize, pressure=pressure, $
generations=generations, $
interval_wavelength=interval_wavelength, $
redshift_initial=redshift_initial, $
redshift_tolerance=redshift_tolerance, $
fwhm_initial=fwhm_initial, $
fwhm_tolerance=fwhm_tolerance, $
fwhm_min=fwhm_min, fwhm_max=fwhm_max, $
image_output_path=image_output_path, output_path=output_path)
output_filename=output_path+'line_list'
mgfit_save_lines, emissionlines, output_filenameAlternatively, you could load the mgfit object class, which automatically loads the line list database as follows:
mg=obj_new('mgfit')
mg->set_output_path, output_path
mg->set_image_output_path, image_output_path
mg->read_ascii, input_file, wavel, flux
emissionlines = mg->detect_lines(wavel, flux, $
popsize=popsize, pressure=pressure, $
generations=generations, $
interval_wavelength=interval_wavelength, $
redshift_initial=redshift_initial, $
redshift_tolerance=redshift_tolerance, $
fwhm_initial=fwhm_initial, $
fwhm_tolerance=fwhm_tolerance, $
fwhm_min=fwhm_min, fwhm_max=fwhm_max)
output_filename=output_path+'line_list'
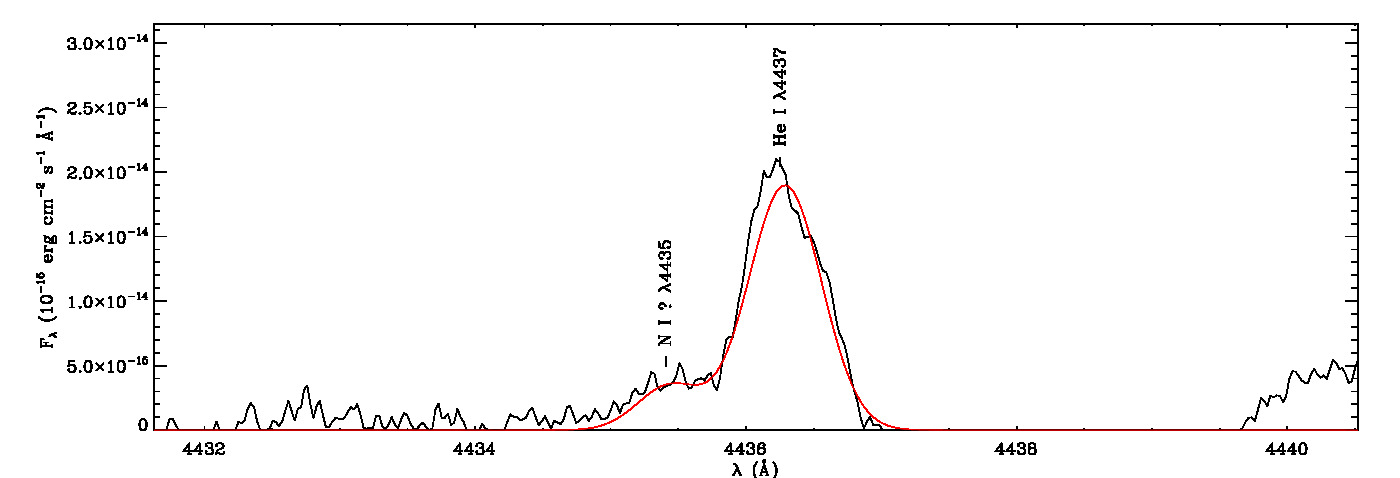
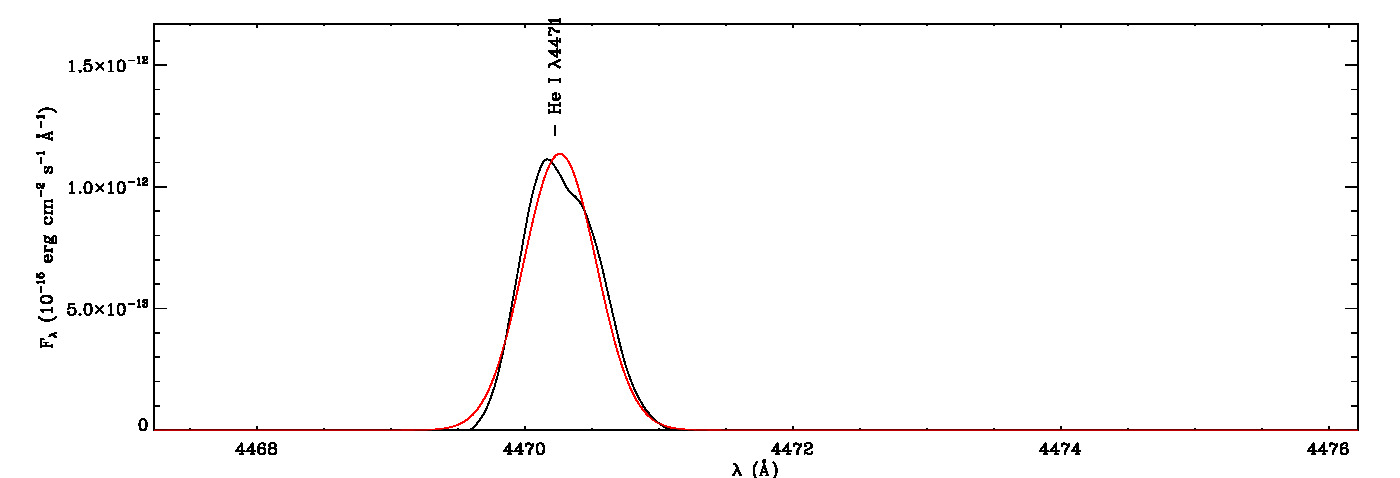
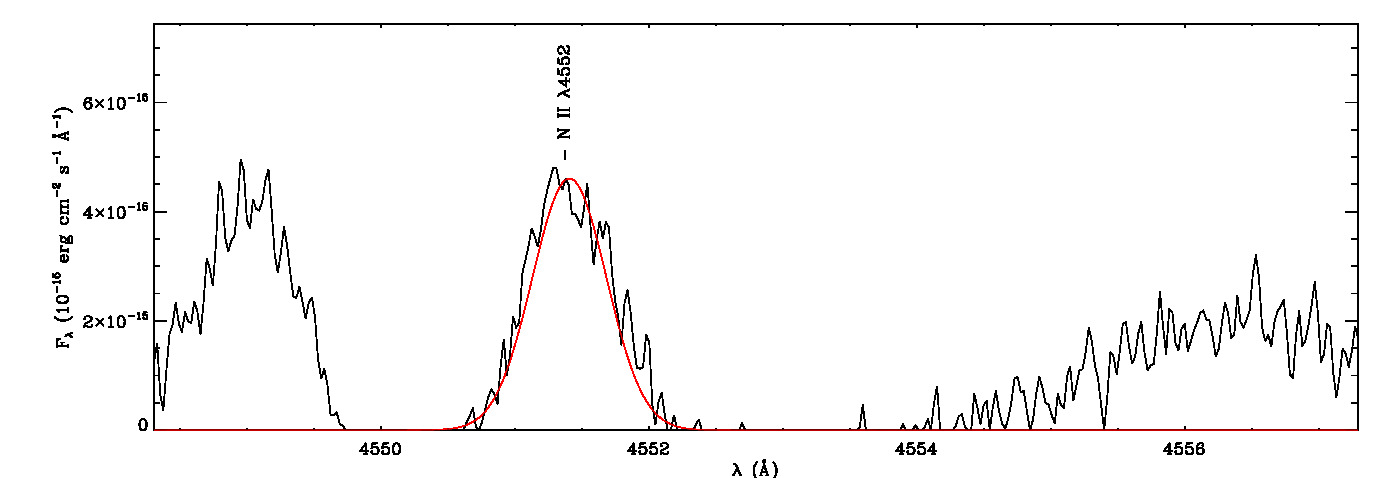
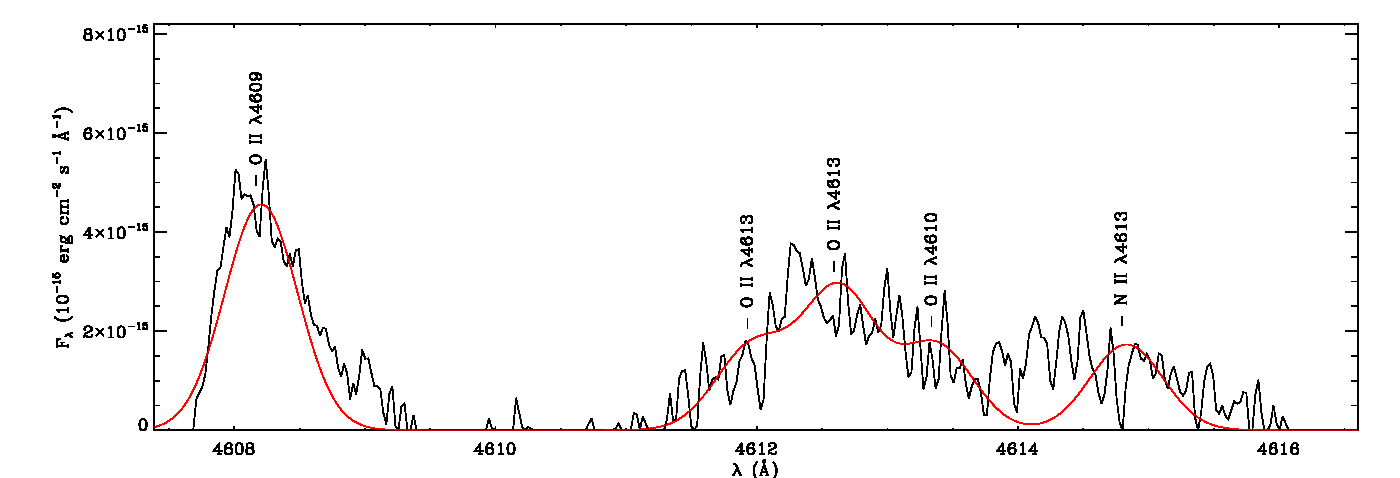
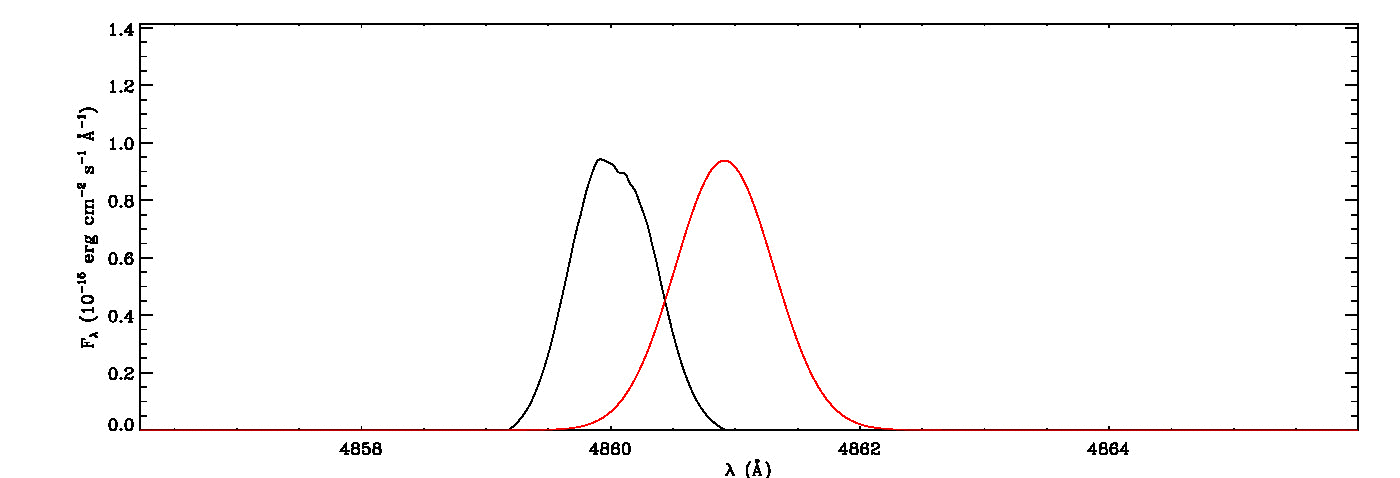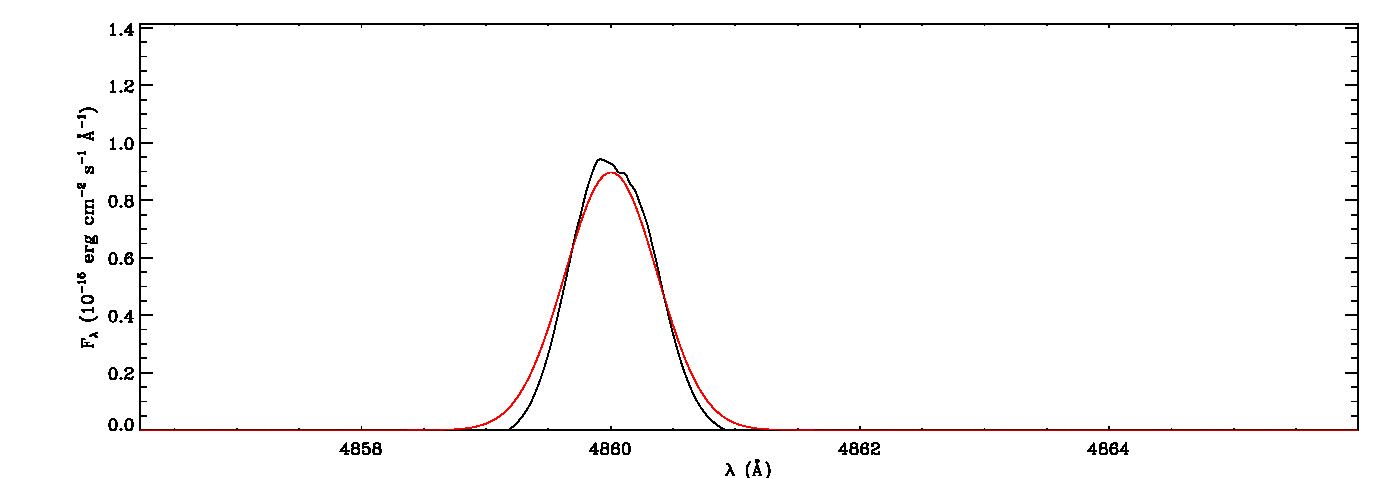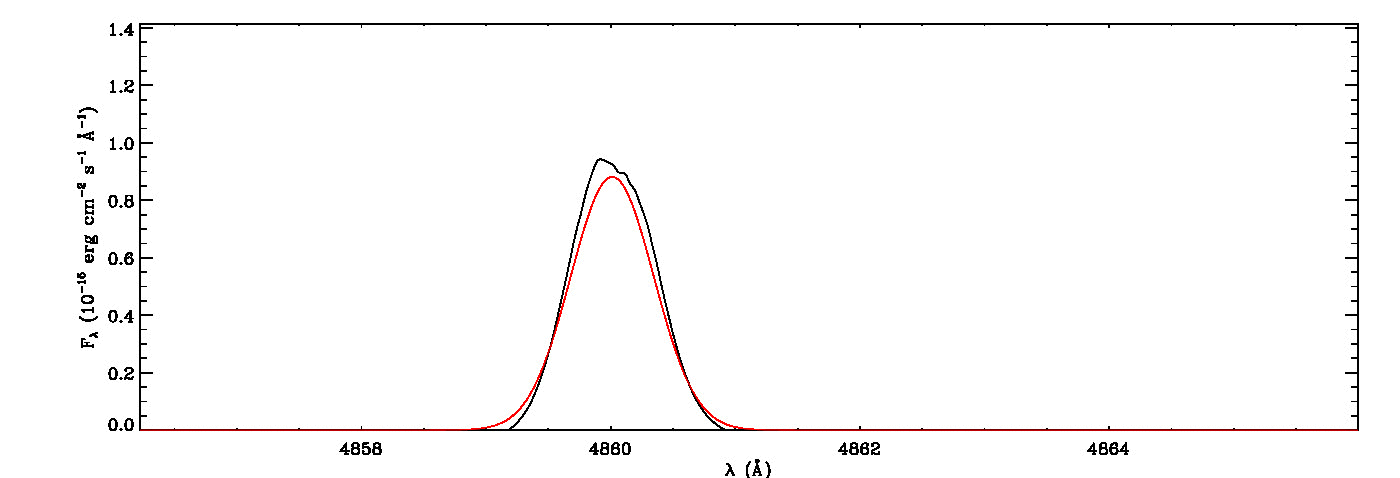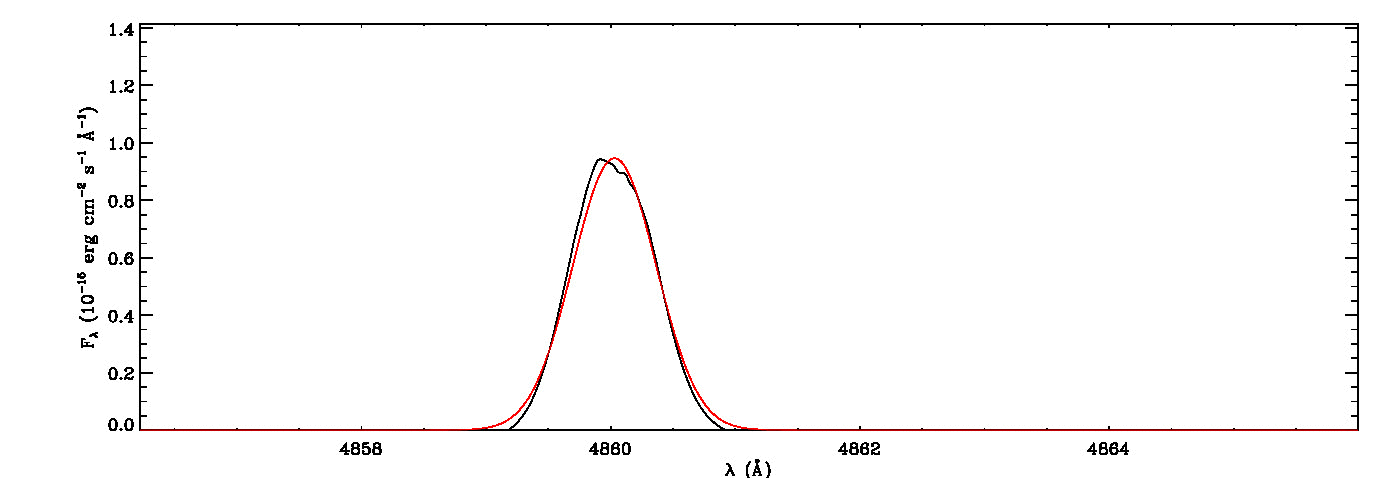
mg->save_lines, emissionlines, output_filenameIt will take a while to identify lines and fit Gaussian curves. You need to check the images of fitted lines stored in the image folder to remove some misidentified lines manually from the final list.
- To get better results, you should use a higher number of generations or/and a higher number of populations, which will increase the computational time, but will result in better fitted lines.
- You need to adjust the FWHM parameters according to the spectral resolution of your observations.
- You need to change the redshift parameters for high redshift sources.
For more information on how to use the API functions from the MGFIT-idl libray, please read the API Documentation published on mgfit.github.io/MGFIT-idl.
| Documentation | https://mgfit.github.io/MGFIT-idl/doc/ |
| Repository | https://github.com/mgfit/MGFIT-idl |
| Issues & Ideas | https://github.com/mgfit/MGFIT-idl/issues |
| Archive | 10.5281/zenodo.4495916 |