-
Notifications
You must be signed in to change notification settings - Fork 131
Tips for finishing genomes
Ideally, a Unicycler hybrid assembly will result in a completed bacterial genome all by itself. But if it doesn't, then the genome might need 'manual completion'. This page contains some tips and tricks to help you along.
In a completed assembly, each chromosome/plasmid in the genome is represented by a single contig. How do you tell if a Unicycler assembly is complete? Unicycler's output/log might help. In the 'Bridged assembly graph' section towards the end of Unicycler's pipeline, it will summarise the graph components:
Component Segments Links Length N50 Longest segment Status
total 7 7 5,676,472 5,583,468 5,583,468
1 1 1 5,583,468 5,583,468 5,583,468 complete
2 1 1 71,104 71,104 71,104 complete
3 1 1 6,657 6,657 6,657 complete
4 1 1 5,783 5,783 5,783 complete
5 1 1 3,514 3,514 3,514 complete
6 1 1 3,223 3,223 3,223 complete
7 1 1 2,723 2,723 2,723 complete
Unicycler considers a component complete if it is circular: one segment and one link. This doesn't quite apply if your bacterial genome has linear chromosomes/plasmids, in which case a complete component would have no links.
You could also view the assembly graph (assembly.gfa) in Bandage and check that each contig is circular:
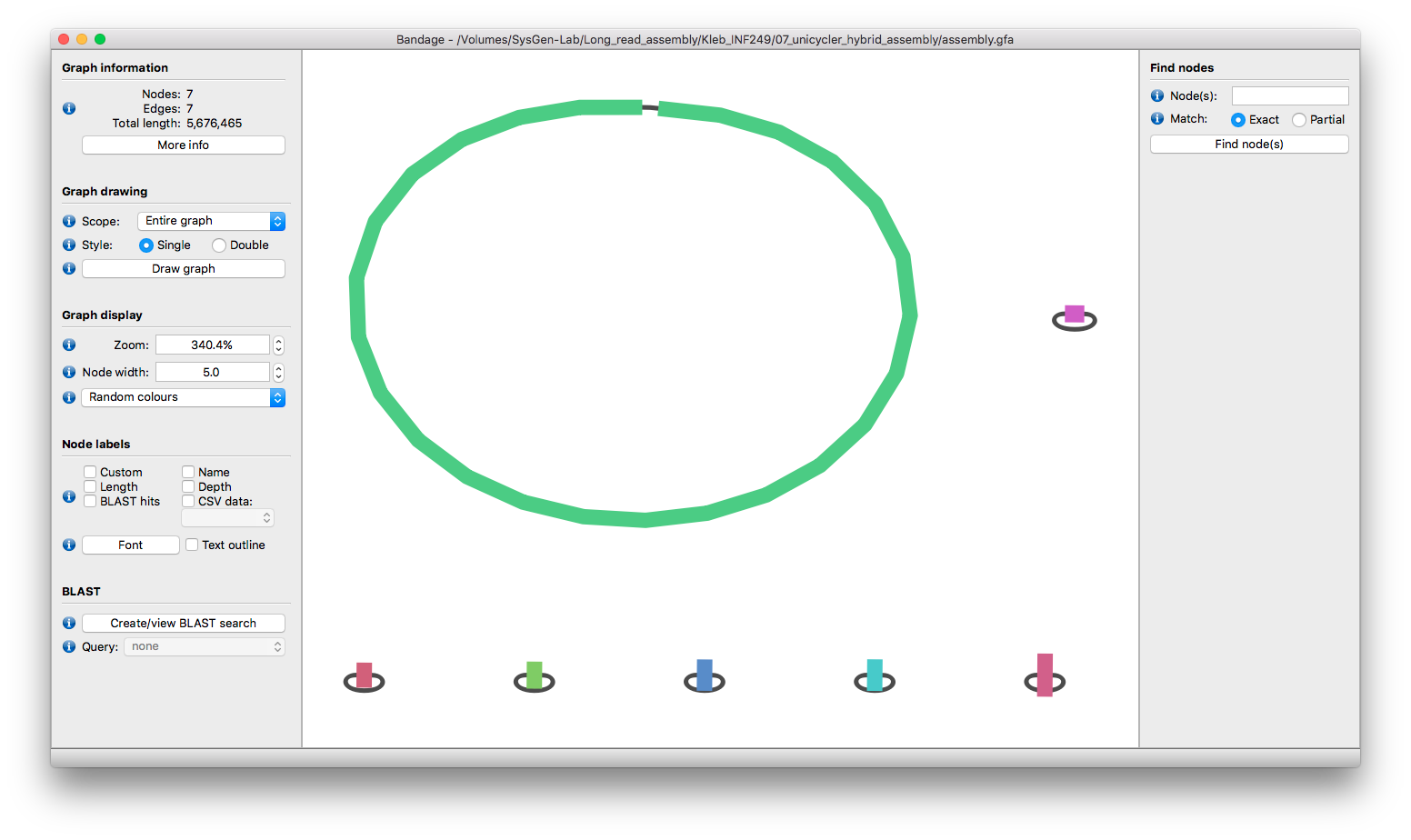 If that's what your graph looks like, then Unicycler completed the assembly on its own!
If that's what your graph looks like, then Unicycler completed the assembly on its own!
But what if it's not complete? The Unicycler log might have something like this:
Component Segments Links Length N50 Longest segment Status
total 23 29 5,819,363 5,242,094 5,242,094
1 1 1 5,242,094 5,242,094 5,242,094 complete
2 1 1 252,269 252,269 252,269 complete
3 1 1 130,933 130,933 130,933 complete
4 1 1 110,494 110,494 110,494 complete
5 1 1 69,826 69,826 69,826 complete
6 1 1 5,783 5,783 5,783 complete
7 17 23 7,964 1,023 3,382 incomplete
and the Bandage graph might look like this:
 Yuck! This genome needs some manual completion...
Yuck! This genome needs some manual completion...
There are many reasons why Unicycler might fail to complete a hybrid assembly, and so there is no single easy method for manual completion. You'll need to rely on detective work and bioinformatics-know-how. Some general methods which may help are:
- Using Bandage to visualise the assembly graphs from various stages of the Unicycler pipeline.
- Gathering long reads for incomplete regions of the assembly (see Read extraction) and BLASTing them to the graphs.
- Aligning short and/or long reads to the assembly and examining the alignments in IGV or Artemis.
- Using other assemblers (e.g. Canu) on the reads and comparing the results to Unicycler's assembly.
Helpful software:
To get you going, here are some real-world examples of assemblies which failed to complete and how I went about manual completion: