Microbiome analysis of ten patient fecal samples for the identification of diet-derived metabolites and their effects on mental health.
BINF 6999 Project for Summer 2022
Van Raay Lab at the University of Guelph
Primary Advisor: Dr. Terry Van Raay
Secondary Advisor: Dr. Lewis Lukens
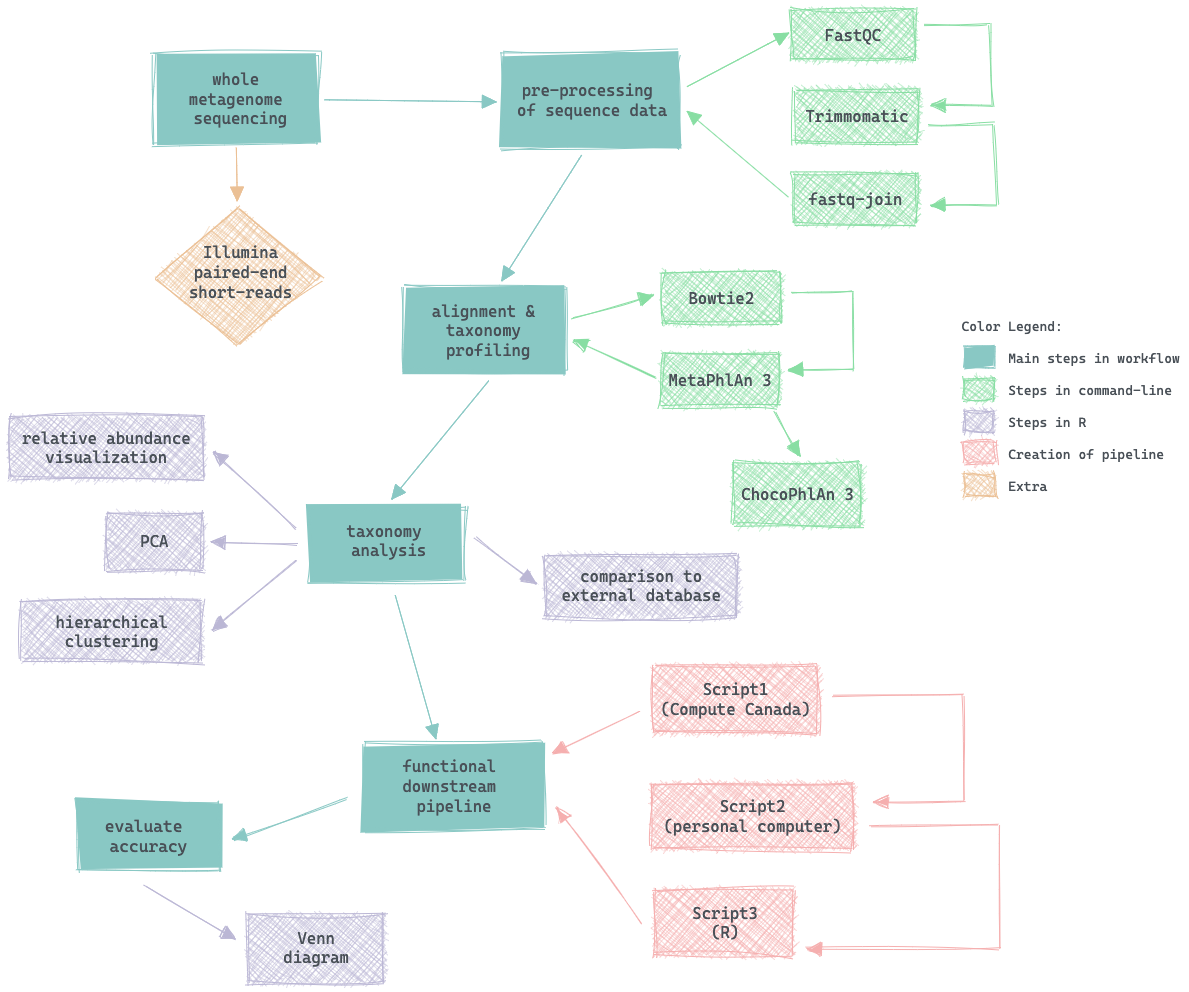
Whole metagenome sequences were used from ten patient fecal samples. These sequences were pre-processed in command-line and the steps can be viewed in the Analysis_Log.txt file.
Diet is a key factor contributing to the balance of the gut microbiome (Berding et al., 2021). It has been proven to be a strong component in the regulation of the brain-gut axis; in particular, the specific metabolites produced by gut microbiota have a considerable impact on the brain health of the individual (Berding et al., 2021). However, it has been difficult for researchers to assess a diet’s precise impacts upon mental health, as other factors such as the individuals’ genetics, age, race, weight, and lifestyle habits must also be taken into consideration. The main gap in knowledge arises from understanding the link between the type of diet and its effects on the developing nervous system. This project serves as an initial stepping-stone upon which changes in neurodevelopmental processes and gene expression may be evaluated, with respect to four specific diets of interest.
The primary goal of this project is to carry out data analysis to direct decision making. Two donor samples representative of the group will be identified based on microbiome analysis of nine neurotypical children’s fecal samples. This selection will be based upon a combination of bioinformatic analyses such as statistical evaluation of relative taxonomic abundance, principal component analysis and hierarchical clustering (Figure 1). The crucial feature of this selection is to prevent the use of an outlier in downstream analysis, where the chosen fecal samples will be inoculated into defined RoboGut (RG) simulated gut environments (McDonald et al., 2013) against four diets of interest. The secondary objective is to create a functional and reproducible pipeline for the future analysis of these RG samples. The whole metagenome sequencing (WMS) file will serve as input, akin to the primary goal, and the output will be the sample’s relative taxonomic abundance. This is important, as the relative abundance allows the Van Raay Lab to evaluate the differences in microbiome among and between samples. There are several software available to carry out this proposed workflow, such as Bracken 2.5, mOTUs2 and MetaPhlAn 3 (Beghini et al., 2021; Lu et al., 2017; Milanese et al., 2019). The bioBakery tools (Beghini et al., 2021) have been selected after assessing their benchmark analysis; the latter, MetaPhlAn 3, yields a greater F1 score for species-level precision relative to the former. In addition, the computational power utilized by MetaPhlAn 3 is very low in comparison (Beghini et al., 2021), and can be used without a server. Figure 1 shows an overview of the project workflow along with the key software employed to carry out analyses. The creation of the downstream pipeline will allow for reproducibility from the initial acquisition of raw reads to the relative abundance output, which includes pre-processing steps such as quality control and sequence trimming. This pipeline will be distinct from pre-existing scripts, as it will be uniquely created for the Van Raay Lab’s RG analyses encompassing the use of various established software with low computational power, as visualized in Figure 1. This customized pipeline will also be evaluated using standard and/or engineered samples to confirm its accuracy.
The foundation of this project is important, both to science and society, as there are distinct correlations between diet and mental health, yet minimal research has been conducted on the causative factors of the involved diet-derived metabolites. Identification of the two donor samples is the initial step to be able to answer this dilemma and the use of the new pipeline will allow for consistency over the course of the project. The ultimate goal is to identify the specific dietary fibres which contribute to beneficial impacts upon mental well-being.