Release v1.0.4
This release of JBrowse Web includes a great many small improvements and bug fixes, see the full changelog below.
Some particularly salient improvements include:
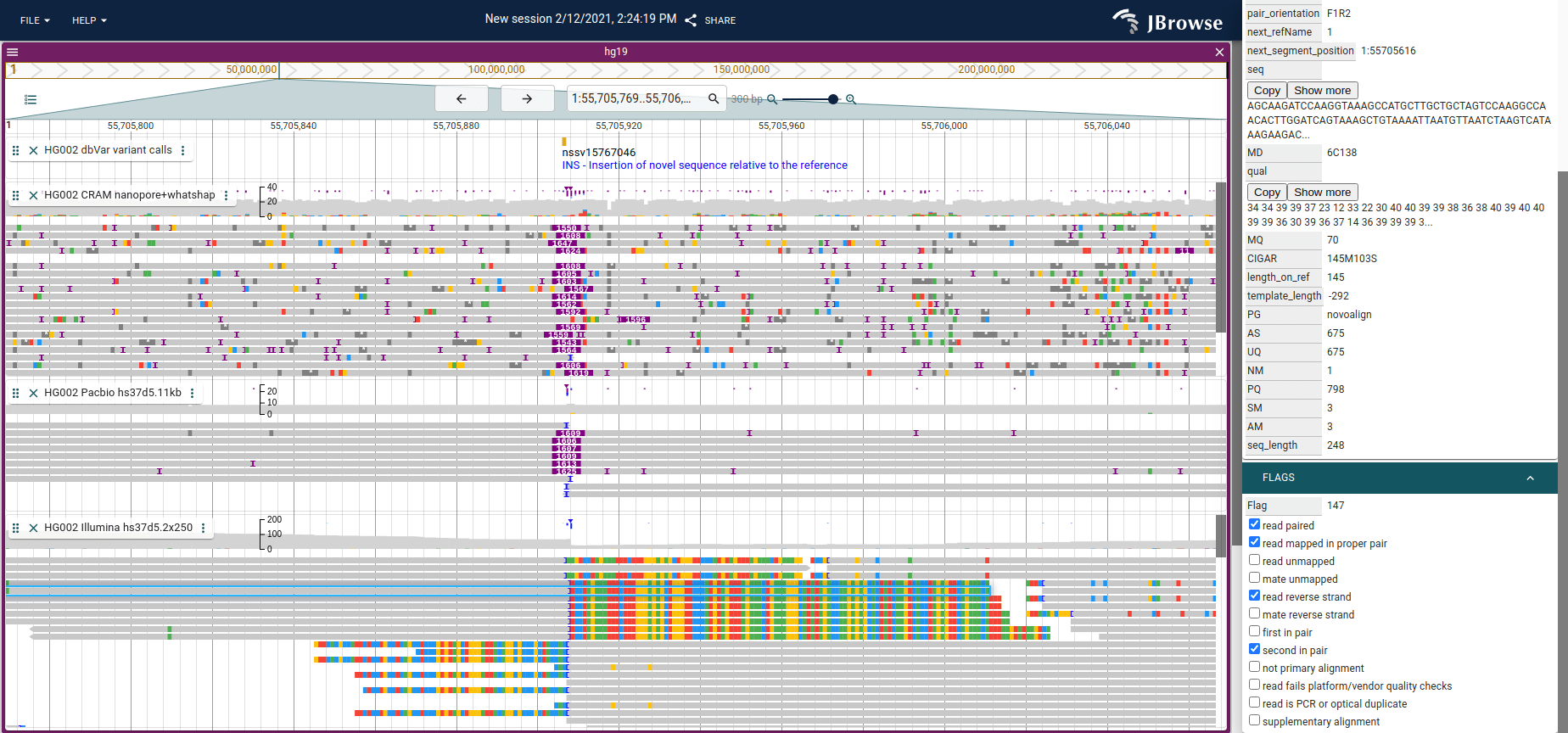
Better indications for insertions
The alignments track received a couple updates including "large insertion indicators" for large indels, and also an upside-down count of clipping or insertion events. There is also a triangular indicator plotted when the insertion/clip count exceeds a threshold at that position defaulted to 30% of reads
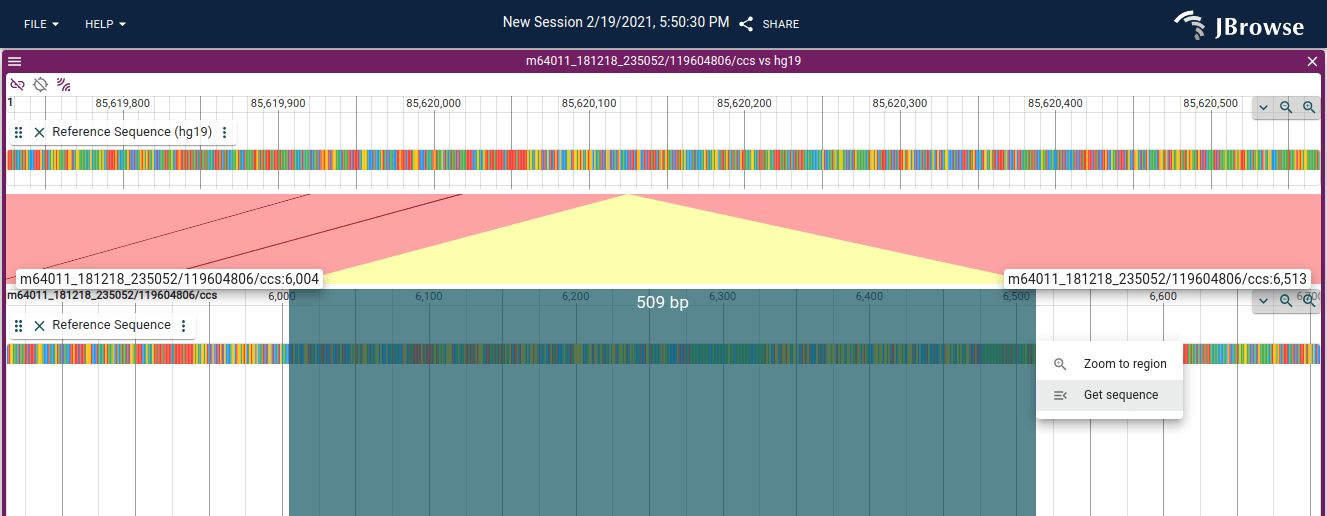
Click and drag the overview bar to "Get sequence"
Users can now download regions of sequence by selecting a region in the linear genome view and clicking "get sequence". See the demonstration video below:
You can also "get sequence" in the read vs reference view, which allows you to "get sequence" for the inserted bases or softclipped bases from a read alignment
A long-requested feature, implemented in #1588 by @teresam856!
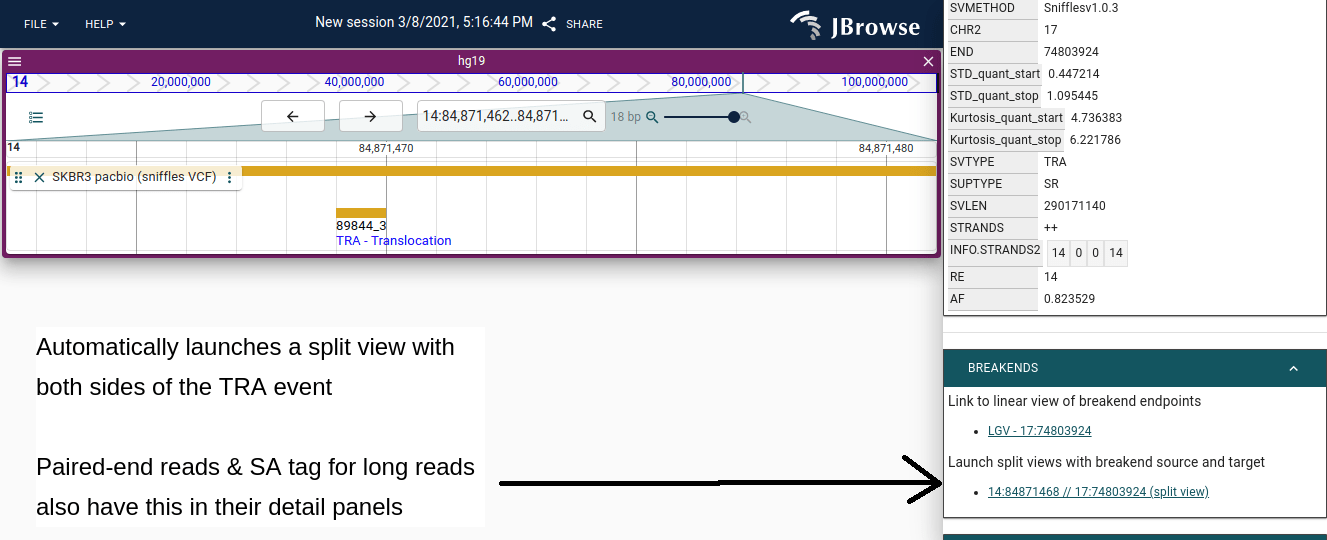
Enhanced navigation of paired end reads and BND/TRA breakends
Feature detail panels for BND/TRA features, long split-alignments, and paired-end reads have links to navigate or popup of the breakpoint split views with their mates.
Implemented by @cmdcolin in #1701.
🚀 Enhancement
core- #1758 Add ability to get stitched together CDS, protein, and cDNA sequences in feature details (@cmdcolin)
- #1721 Manually adjust feature height and spacing on alignments track (@cmdcolin)
- #1728 Add list of loaded plugins to the "About widget" (@rbuels)
- #1711 Add plugin top-level configuration (@teresam856)
- #1699 Add sequence track for both read and reference genome in the "Linear read vs ref" comparison (@cmdcolin)
- #1701 Add clickable navigation links to supplementary alignments/paired ends locations and BND/TRA endpoints in detail widgets (@cmdcolin)
- #1601 Add ability to color by per-base quality in alignment tracks (@cmdcolin)
- #1640 Move stats calculation to BaseFeatureAdapter (@cmdcolin)
- #1588 Add "Get sequence" action to LGV rubber-band (@teresam856)
- Other
- #1743 Add color picker and choice of summary score style for wiggle track (@cmdcolin)
- #1763 Add a "CSS reset" to jbrowse-react-linear-genome-view to prevent parent styles from outside the component leaking in (@cmdcolin)
- #1756 Split alignments track menu items into "Pileup" and "SNPCoverage" submenus (@cmdcolin)
- #1742 Add ability to display crosshatches on the wiggle line/xyplot renderer (@cmdcolin)
- #1736 Fix CLI add-track --load inPlace to put exact contents into the config, add better CLI example docs (@cmdcolin)
- #1394 Add new menu items for show/hide feature labels, set max height, and set compact display mode (@cmdcolin)
- #1720 Standardize phred qual scaling between BAM and CRAM and add option to make mismatches render in a lighter color when quality is low (@cmdcolin)
- #1704 Add "Show all regions in assembly" to import form and make import form show entire region when refName selected (@cmdcolin)
- #1687 Threshold for indicators on SNPCoverage + inverted bargraph of interbase counts for sub-threshold events (@cmdcolin)
- #1695 Improve zoomed-out display of quantitative displays tracks when bicolor pivot is active (@cmdcolin)
- #1680 Add on click functionality to quantitative track features (@teresam856)
- #1630 Get column names from BED tabix files and other utils for external jbrowse-plugin-gwas support (@cmdcolin)
- #1709 Improve sorting and filtering in variant detail widget (@cmdcolin)
- #1688 Bold insertion indicator for large insertions on pileup track (@cmdcolin)
- #1669 Allow plain json encoding of the session in the URL (@cmdcolin)
- #1642 Enable locstring navigation from LGV import form (@teresam856)
- #1655 Add GFF3Tabix and BEDTabix inference to JB1 connection (@garrettjstevens)
- #1643 Add an offset that allows all wiggle y-scalebar labels to be visible (@cmdcolin)
- #1632 Displays warnings when receiving a session with custom callbacks (@peterkxie)
- #1615 Increase pileup maxHeight (@cmdcolin)
- #1624 GCContent adapter (@cmdcolin)
- #1614 Add insertion and clip indicators to SNPCoverage views (part of Alignments tracks) (@cmdcolin)
- #1610 Display error message from dynamodb session sharing error (@cmdcolin)
🐛 Bug Fix
- Other
- #1777 Quick fix for block error (@cmdcolin)
- #1748 External plugins load after confirming config warning (@peterkxie)
- #1750 Fix pileup sorting when using string tag (@cmdcolin)
- #1747 Fix the position of the popup menu after rubberband select when there is a margin on the component e.g. in embedded (@cmdcolin)
- #1736 Fix CLI add-track --load inPlace to put exact contents into the config, add better CLI example docs (@cmdcolin)
- #1731 Fix alignment track ability to remember the height of the SNPCoverage subtrack on refresh (@cmdcolin)
- #1719 Fix for navigation past end of chromosome (@cmdcolin)
- #1698 Fix rendering read vs ref comparisons with CIGAR strings that use = sign matches (@cmdcolin)
- #1697 Fix softclipping configuration setting causing bases to be missed (@cmdcolin)
- #1689 Disable copy/delete menu items for reference sequence track (@teresam856)
- #1682 Fix parsing of BED and BEDPE files with comment header for spreadsheet view (@cmdcolin)
- #1679 Fix issue with using launching the add track widget on views that are not displaying any regions (@teresam856)
- #1642 Enable locstring navigation from LGV import form (@teresam856)
- #1626 Bug Fix: specify assembly in locstring (@teresam856)
- #1619 Fix overview scale polygon not appearing properly in some cases (@cmdcolin)
core
📝 Documentation
- #1725 JBrowseR release (@elliothershberg)
- #1677 Config guide updates (@elliothershberg)
- #1665 Add Nextstrain COVID storybook (@elliothershberg)
- #1670 typo in developer guide docs (@teresam856)
- #1592 Website copy edits (@rbuels)
- #1646 Fix "See code" link in CLI docs (@garrettjstevens)
- #1618 Add whole-genome view and color/sort alignments tutorials to user guide (@cmdcolin)
🏠 Internal
- Other
- #1666 Move "mouseover" config from BaseLinearDisplay to LinearBasicDisplay display (@garrettjstevens)
- #1751 Make the variant display derive from the feature display (@cmdcolin)
- #1716 Stringify labels before adding to rendering to avoid undefineds on label.length (@cmdcolin)
- #1713 Add console.error log in block setError (@cmdcolin)
- #1663 Make LGV "initialized" not depend on displayedRegions (@garrettjstevens)
- #1672 Fix import forms crashing if there are no assemblies (@cmdcolin)
- #1644 Bump electron from 9.3.1 to 9.4.0 (@dependabot[bot])
- #1641 Remove codecov pr annotations (@cmdcolin)
- #1609 Add extra checks for release script (@peterkxie)
core- #1762 Add requestidlecallback ponyfill in @jbrowse/core/util (@cmdcolin)
- #1629 Add RegionsAdapter/SequenceAdapter, reorganize base adapters (@garrettjstevens)
- #1625 Make renderArgs consistent and don't duplicate data (@garrettjstevens)
- #1414 Typescriptify and MST'ify the add track workflow (@cmdcolin)