-
Notifications
You must be signed in to change notification settings - Fork 16
3D_Nuclei_Clustering_Tool
Analyze the clustering behavior of nuclei in 3D images. The centers of the nuclei are detected. The nuclei are filtered by the presence of a signal in a different channel. They clustered with the density based algorithm DBSCAN. The nearest neighbor distances between all nuclei and those outside and inside of the clusters are calculated.
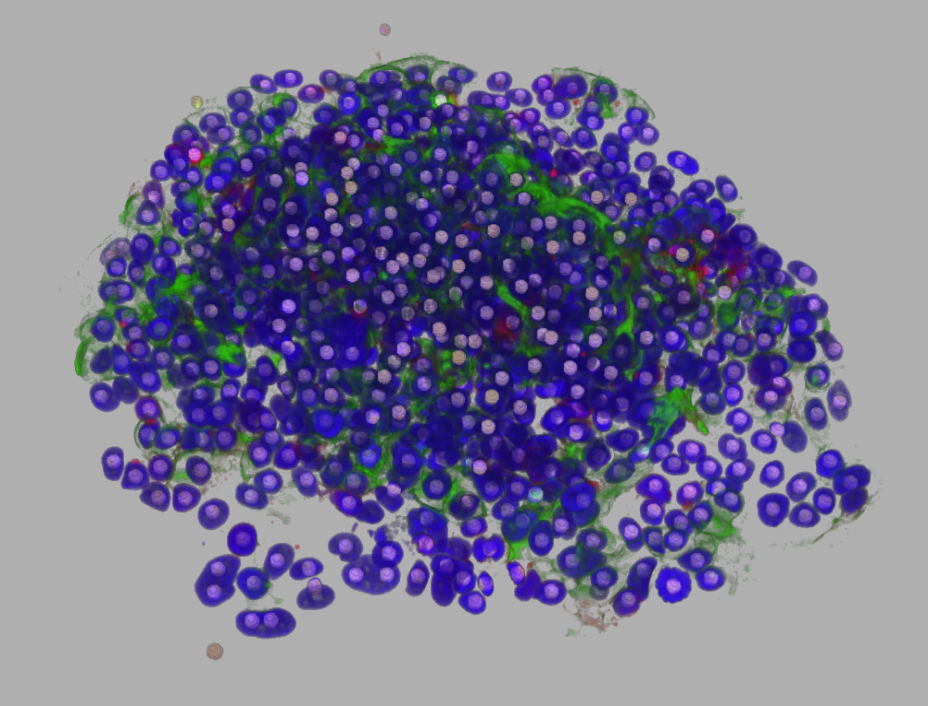
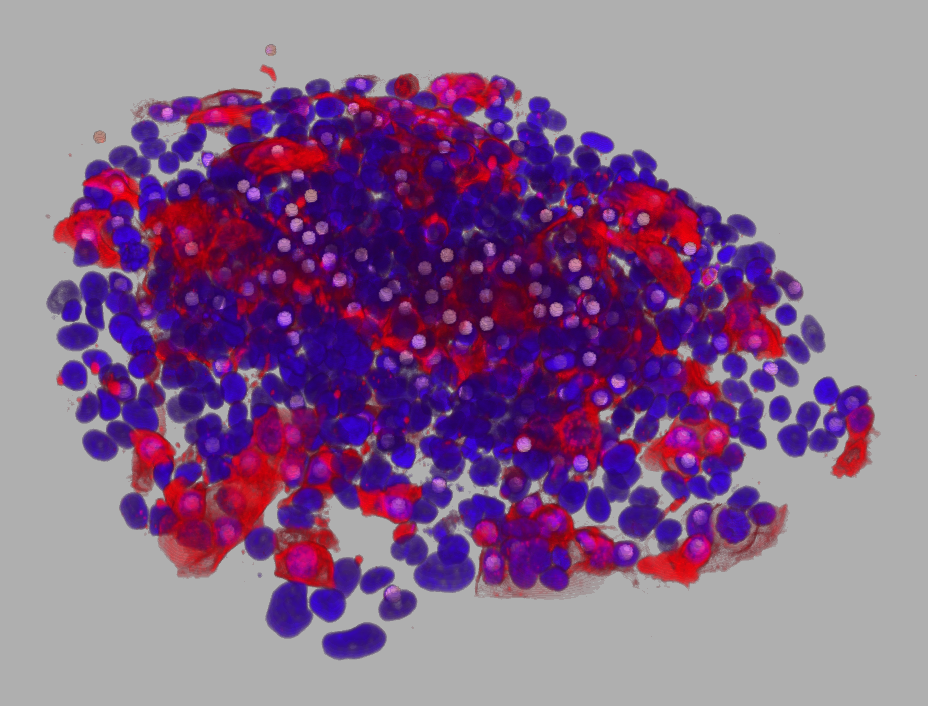


You can download an example data-set:
The source code in git-hub can be found here.
You must have the 3D ImageJ Suite [1] and FeatureJ installed. They are available via the FIJI-update-sites 3D ImageJ Suite and ImageScience.
To install the tool save the three files 3D_nuclei_clustering.ijm, dbscan_clustering_3D.py and nearest_neighbor_distances_3D.py into the folder macros/toolsets of your FIJI installation.
Select the "3D_nuclei_clustering" toolset from the >> button of the ImageJ launcher.
- the first button (the one with the image) opens this help page
- the
p-button runs the complete analysis on the active image - the
b-button runs the batch analysis on all images in a folder
The remaining buttons execute one of the steps of the analysis at a time. This can be helpful for finding the best option-values.
- the
d-button detects the nuclei in the image - the
f-button filters the nuclei, so that only nuclei with an intensity above a threshold in another channel remain. - the
c-button runs the cluster analysis. - the
n-button calculates the nearest-neighbor distances for the active table - the
v-button visualizes the nearest-neighbor connections
[1] Ollion, J., Cochennec, J., Loll, F., Escudé, C., and Boudier, T. (2013). TANGO: a generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics 29, 1840–1841.

 Volker Bäcker
Volker Bäcker