This repository contains a program to generate microstructures using a site-saturation condition and simulate grain growth using the Monte Carlo Potts Model. The generated microstructures can be used as initial conditions for the grain growth simulation, which can be used to study the evolution of grain boundaries over time.
micro_w600_c2_mhalton_o1500_t00010.mp4
-
Download repository:
Without Git:
curl.exe -LJO https://github.com/Neelfrost/microstructure-mas/archive/refs/heads/main.zip; Expand-Archive -Force .\microstructure-mas-main.zip .; cd .\microstructure-mas-mainWith Git:
git clone https://github.com/Neelfrost/microstructure-mas; cd .\microstructure-mas -
Install using pip:
pip install .
mmas.exe --help
usage: mmas [-h] [-w int] [-c int] [-o int] [-m {pseudo,sobol,halton,latin}] [-T float] [-b float] [-g float]
[--simulate] [--color] [--snapshot int] [--save] [--load str]
Microstructure Modeling and Simulation. Generate microstructures using site-saturation condition, and simulate grain
growth using Monte Carlo Potts Model.
options:
-h, --help Show this message and exit.
-w, --width Set the application window width. (default: 500)
-c, --cell-size Define the grid cell size. Lower values result in sharper boundaries. (default: 5, recommended range: 1-10)
-o, --orientations Specify the initial number of grains. Higher values produce smaller grains. (default: 100)
-m, --method Choose the seed generation algorithm. Allowed values are: pseudo, sobol, halton, latin. (default: halton)
-T, --temperature Set the simulation temperature. Higher values increase the likelihood of unfavorable grain boundary migration. (default: 0, recommended range: 0-2)
-b, --boltz Specify the Boltzmann constant. (default: 1)
-g, --grain Set the grain boundary energy. (default: 1)
--simulate Enable grain growth simulation. (default: false)
--color Display grains in color instead of grayscale. (default: false)
--snapshot Save snapshots of the microstructure at specified intervals (in seconds). Without simulation, only one snapshot is saved. (default: never)
--save Save microstructure data to a file. (default: false)
--load Load microstructure data from a file. This option can override or be combined with other options like --temperature, --grain, --boltz, --simulate,
--color, and --snapshot.
-hb, --highlight-boundaries
Process snapshots of a microstructure from a specified folder to extract and display only grain boundaries. The processed snapshots are saved with
highlighted grain boundaries, removing the original colored grain representation.Note: This requires imagemagick (https://imagemagick.org) to be
installed.
The program parameters, such as the number of grains, and the temperature of the Potts Model, can be adjusted as per above.
| Pseudo | Sobol |
|---|---|
 |
 |
| Halton | Latin Hypercube |
|---|---|
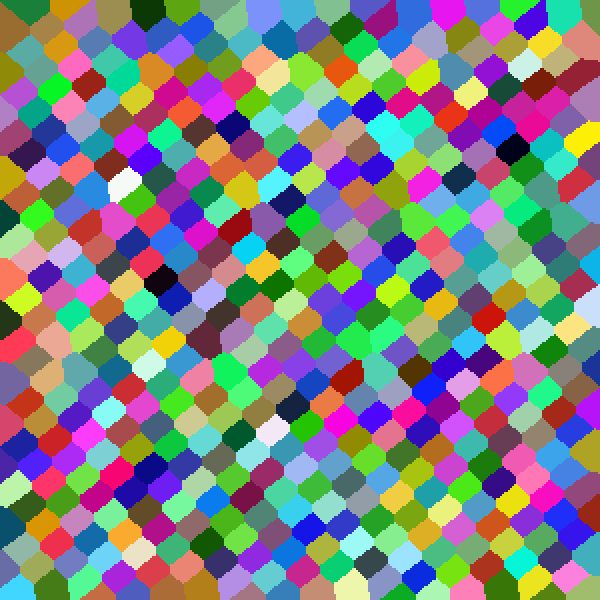 |
 |
The program is intended to provide a qualitative understanding of the grain growth process, and is not intended to be used for quantitative predictions or engineering applications.
- Paulo Blikstein, André Paulo Tschiptschin, Monte Carlo Simulation of Grain Growth.
- S. Sista And T. Debroy, Three-Dimensional Monte Carlo Simulation of Grain Growth in Zone-Refined Iron.
- N. Maazi, Conversion of Monte Carlo Steps to Real Time for Grain Growth Simulation.