-
Notifications
You must be signed in to change notification settings - Fork 4
1000 Genomes example
fgardos edited this page Feb 25, 2014
·
19 revisions
BiERapp includes an example based on 1000 Genomes data.
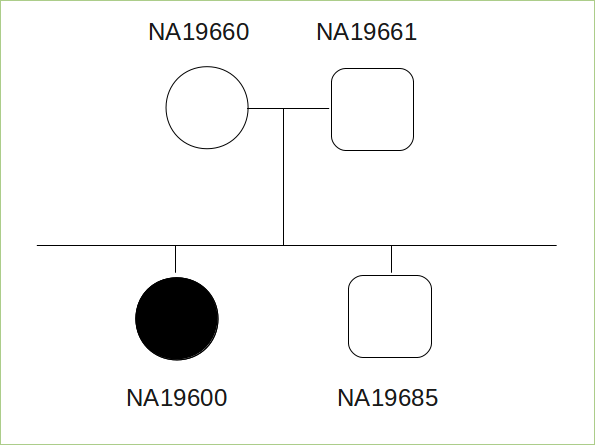
- We generate a synthetic family consisting of 4 people: father (id: NA19661), mother (id: NA19660), son (id: NA19685) and daughter (id: NA19600). The first 3 people come from m008 trio in 1000 Genomes data.
- It was created a VCF file for daughter including variants of X chromosome from the mother and the rest of chromosomes, the father and mother provided randomly 50% of the variants.
- A multi-sample VCF file was prepared and 1000 variants of the family were randomly selected.
- Mother, father and son are unaffected. Daughter is affected, so it was included a mutation for her: TC / C in the position chr11: 61724448 (this deletion is causing a frame-shift).
A multi-sample VCF file. (When having only one sample the input is an individual VCF file).
- Uploading multi-sample VCF file in BiERapp.
- After visualizing initial results you can use different filtering options to refine your variant selection strategy.
For example, we want to detect recessive variants (mother, father and son are unaffected; daughter is affected) in the chromosome 11, so these are the filtering options:
- Region. We include the chromosome of interest: 11.
- Segregation. Here we define the genotype for each sample: father (NA19661), mother (NA19660) and son ( NA19685) are 0/1; daughter (NA19600) is 1/1. BiERapp detects the simulated mutation previously (TC / C in the position chr11: 61724448) and describe its effects. From Genome Viewer is possible to visualize detected variants.
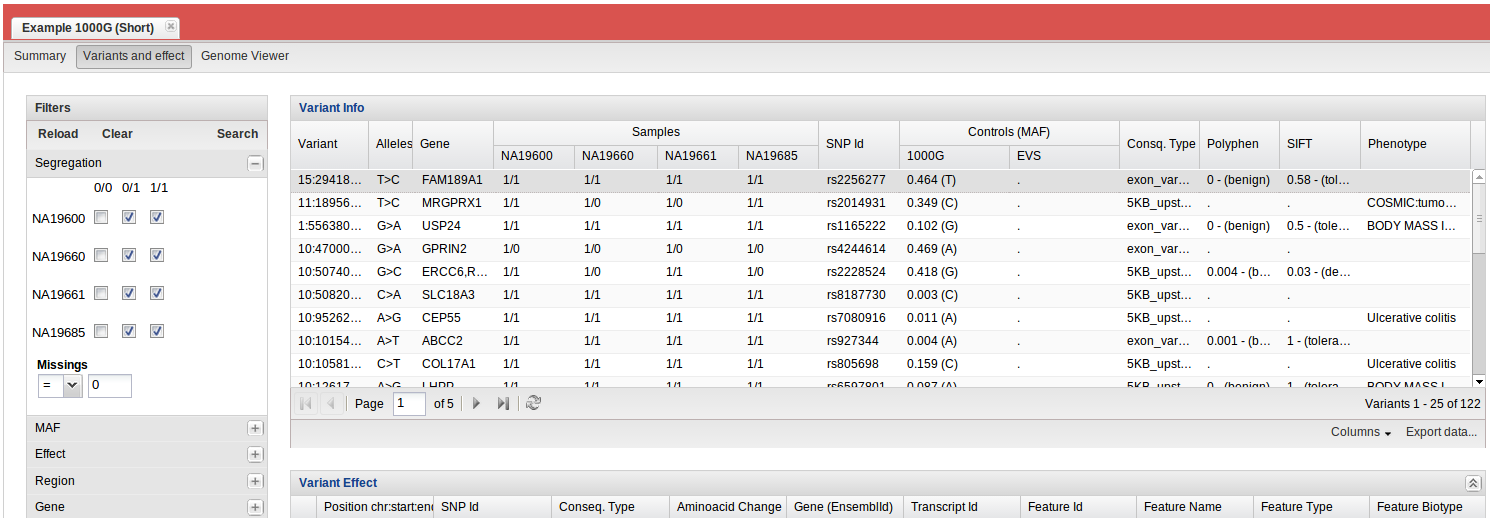
- Summary: global statistics and a graphical representation for consequence type.
- Variants and effects: detected variants and its effects for all samples.
- Genome viewer: selected variants can be visualized from this genome browser