Version 1.46.1
Version 1.46.1 introduces a new SNPsites plugin.
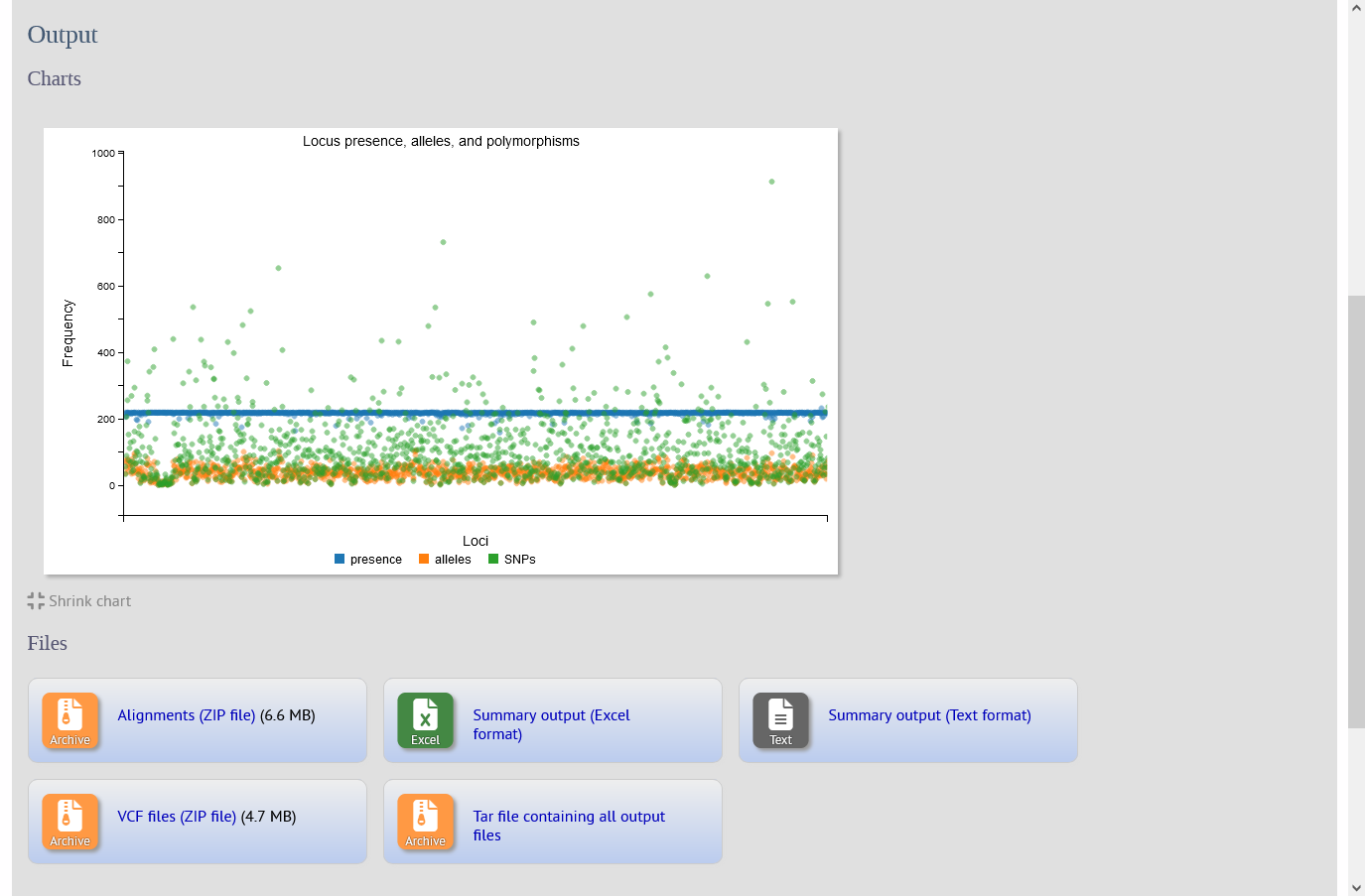
The SNPsites plugin aligns sequences for specified loci for an isolate dataset. The alignment is then passed to snp-sites to identify SNP positions. Output consists of a summary table including the number of alleles and polymorphic sites found for each locus, an interactive D3 chart that displays the summary, and ZIP files containing alignment FASTAs and VCF files for each locus.
Note that snp-sites needs to be installed and its path set in bigsdb.conf, e.g.
#snp-sites##########
snp_sites_path=/usr/bin/snp-sites
Full Changelog: v_1.46.0...v_1.46.1