Releases: GMOD/jbrowse-components
Release v1.3.3
This is a small bugfix release that includes notable fixes for the
methylation/modifications drawing (the negative strand reads displayed the data
incorrectly) and the reference name selector
Please check the release notes for more details!
1.3.3 (2021-08-02)
Packages in this release
🚀 Enhancement
- Other
core
🐛 Bug Fix
- Other
- #2159 Stop local storage quota-exceeded errors preventing the app from starting (@cmdcolin)
- #2161 Remove outline from clicking on SVG chord tracks (@cmdcolin)
- #2157 Fix rendering of negative strand alignment modifications/methylation (@cmdcolin)
- #2131 Fix mouseovers/click handlers after "force load" button pressed (@cmdcolin)
- #2128 Fix using the "Color by modifications" setting on files that need ref renaming (@cmdcolin)
- #2115 Fix bug where sometimes plugin could not be removed from UI (@garrettjstevens)
- #2119 Fix loading indicator on the reference sequence selector getting stuck (@cmdcolin)
core- #2101 Fix behavior of the end-of-list indicator in refNameAutocomplete to always display as a disabled item (@teresam856)
🏠 Internal
- Other
- #2152 Remove storybook symlink workaround (@garrettjstevens)
core
Committers: 3
- Colin Diesh (@cmdcolin)
- Garrett Stevens (@garrettjstevens)
- Teresa Martinez (@teresam856)
Release v1.3.2
This is a hotfix release that fixes a bug that prevented the
@jbrowse/react-linear-genome-view component from being used in v1.3.1
It also contains a couple other small improvements
1.3.2 (2021-07-07)
Packages in this release
| Package | Download |
|---|---|
| @jbrowse/core | https://www.npmjs.com/package/@jbrowse/core |
| @jbrowse/plugin-circular-view | https://www.npmjs.com/package/@jbrowse/plugin-circular-view |
| @jbrowse/plugin-spreadsheet-view | |
| @jbrowse/plugin-sv-inspector | |
| @jbrowse/plugin-svg | https://www.npmjs.com/package/@jbrowse/plugin-svg |
| @jbrowse/plugin-variants | https://www.npmjs.com/package/@jbrowse/plugin-variants |
| @jbrowse/desktop | |
| @jbrowse/react-linear-genome-view | https://www.npmjs.com/package/@jbrowse/react-linear-genome-view |
| @jbrowse/web |
🚀 Enhancement
- Other
core
🐛 Bug Fix
- #2109 Make sure to wait for assembly to load before downloading canonical refnames in SV inspector (@cmdcolin)
- #2111 Fix "Can't resolve '@jbrowse/plugin-legacy-jbrowse'" in
@jbrowse/react-linear-genome-view(@garrettjstevens)
Committers: 2
- Colin Diesh (@cmdcolin)
- Garrett Stevens (@garrettjstevens)
Release v1.3.1
Hello everyone! This release offers a couple important bug fixes.
For users of @jbrowse/react-linear-genome-view, we have fixes that improve
speed, CSS style consistency, and theming. We also have another speed
improvement for users with many scaffolds or contigs. Please see the release
notes below for more details!
1.3.1 (2021-07-06)
Packages in this release
🚀 Enhancement
core- #2094 More usage of typography to improve consistent text styling (@cmdcolin)
- #2068 Add non-indexed and plaintext VCF Adapter to variants plugin (@carolinebridge-oicr)
- #2067 Better error message if a file location has an empty string (@garrettjstevens)
- #2064 Export offscreenCanvasPonyfil from core/util (@garrettjstevens)
- #2060 Improve performance with large numbers of reference sequences by using MST volatiles (@cmdcolin)
- #2050 Configurable app logo for web (@elliothershberg)
- Other
- #2104 Use ScopedCssBaseline to help style the embedded component (@cmdcolin)
- #2015 Deprecate ThemeProvider in
@jbrowse/react-linear-genome-view(@garrettjstevens)
🐛 Bug Fix
- Other
- #2097 Improve speed for laying out features for embedded/mainthreadrpc scenarios (@cmdcolin)
- #2096 Fix issue with page reload after editing session title (@cmdcolin)
- #2074 Fix support for opening local files in spreadsheet/SV inspector (@cmdcolin)
- #2061 Fix issue with using --force error when no track was previously loaded (@cmdcolin)
- #2024 Flip drawing of negative strand PAF features in linear synteny and dotplot views (@cmdcolin)
- #2023 Fix infinite loop in adding some plugins on desktop (@cmdcolin)
- #2019 Fix session import to use blob map for opening local files (@cmdcolin)
core- #2071 Add indicator to the end of ref name dropdown to suggest user to type the searchbox for more (@teresam856)
- #2056 Fix infinite recursion in FromConfigAdaptor by avoiding mutating the passed in data when using SimpleFeature (@cmdcolin)
- #2018 Fix 3'UTR in sequence detail panels when no UTRs are in gff (@cmdcolin)
🏠 Internal
core- #1967 Omit configurationSchema snapshot when it matches the default (@garrettjstevens)
- #2078 Restore eslint rule for no-unused-vars (@cmdcolin)
- #2051 Add missing named exports to shared core modules (@garrettjstevens)
- #2045 Add basic architecture for text searching (@teresam856)
- Other
Committers: 5
- Caroline Bridge (@carolinebridge-oicr)
- Colin Diesh (@cmdcolin)
- Elliot Hershberg (@elliothershberg)
- Garrett Stevens (@garrettjstevens)
- Teresa Martinez (@teresam856)
Release v1.3.0
We're excited to announce the v1.3.0 release of JBrowse Web! Some highlights of
this release include:
In-app plugin store
Building on the plugin store on our website in the last release, we're now
excited to announce that plugins can be installed from within JBrowse Web!
Plugins from our plugin store can now be installed with the click of a button.
Open local files
JBrowse Web now has the ability for tracks to use files on your local hard
drive. This is a great option if you want to visualize files you have locally
without uploading them to a server. These files will need to be re-opened each
time the app is opened or refreshed, but more robust handling of local files
will be available when we release JBrowse Desktop.
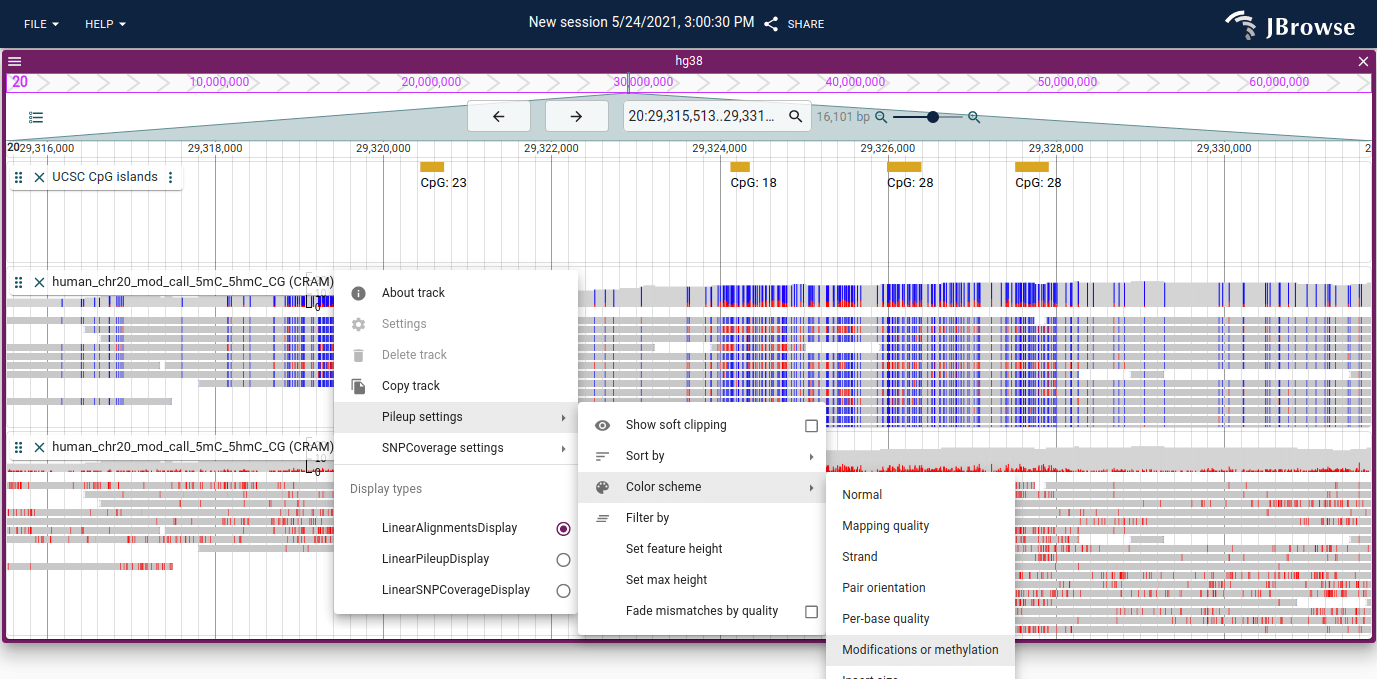
Color by MM and MP/ML tags in BAM/CRAM
The MM and MP/ML tags can be used to color alignments tracks by either base
modifications or by methylation. The modifications mode is exciting because it
can show arbitrary DNA/RNA modifications, and the methylation mode uses
specific CpG context to show both modified and unmodified CpGs.
In this screenshot, the top alignments track is colored by methylation and the
bottom alignments track is colored by base modification.
1.3.0 (2021-05-24)
Packages in this release
| Package | Download |
|---|---|
| @jbrowse/core | https://www.npmjs.com/package/@jbrowse/core |
| @jbrowse/development-tools | https://www.npmjs.com/package/@jbrowse/development-tools |
| @jbrowse/plugin-alignments | https://www.npmjs.com/package/@jbrowse/plugin-alignments |
| @jbrowse/plugin-breakpoint-split-view | |
| @jbrowse/plugin-circular-view | https://www.npmjs.com/package/@jbrowse/plugin-circular-view |
| @jbrowse/plugin-config | https://www.npmjs.com/package/@jbrowse/plugin-config |
| @jbrowse/plugin-data-management | https://www.npmjs.com/package/@jbrowse/plugin-data-management |
| @jbrowse/plugin-dotplot-view | |
| @jbrowse/plugin-filtering | |
| @jbrowse/plugin-gff3 | https://www.npmjs.com/package/@jbrowse/plugin-gff3 |
| @jbrowse/plugin-hic | |
| @jbrowse/plugin-legacy-jbrowse | |
| @jbrowse/plugin-linear-comparative-view | |
| @jbrowse/plugin-linear-genome-view | https://www.npmjs.com/package/@jbrowse/plugin-linear-genome-view |
| @jbrowse/plugin-lollipop | |
| @jbrowse/plugin-menus | |
| @jbrowse/plugin-protein | |
| @jbrowse/plugin-rdf | |
| @jbrowse/plugin-sequence | https://www.npmjs.com/package/@jbrowse/plugin-sequence |
| @jbrowse/plugin-spreadsheet-view | |
| @jbrowse/plugin-sv-inspector | |
| @jbrowse/plugin-svg | https://www.npmjs.com/package/@jbrowse/plugin-svg |
| @jbrowse/plugin-trackhub-registry | |
| @jbrowse/plugin-variants | https://www.npmjs.com/package/@jbrowse/plugin-variants |
| @jbrowse/plugin-wiggle | https://www.npmjs.com/package/@jbrowse/plugin-wiggle |
| @jbrowse/cli | https://www.npmjs.com/package/@jbrowse/cli |
| @jbrowse/desktop | |
| @jbrowse/react-linear-genome-view | https://www.npmjs.com/package/@jbrowse/react-linear-genome-view |
| @jbrowse/web |
🚀 Enhancement
- Other
core- #1982 Allow manually specifying adapter type if filename does not match expected pattern (@cmdcolin)
- #1975 Allow local files on the users computer to be opened as tracks in jbrowse-web (@cmdcolin)
- #1865 Show modified bases using MM and MP/ML tags in BAM/CRAM (@cmdcolin)
- #1984 Better feature details when there are short arrays of json supplied as feature data (@cmdcolin)
- #1931 Create in app graphical plugin store (@elliothershberg)
🐛 Bug Fix
core- #1985 Avoid error calculating UTR on features that have no exon subfeatures (@cmdcolin)
- #1954 Add more environments to configSchema create calls to fix ability to use custom jexl commands with main thread rendering (@cmdcolin)
- #1963 Fix ability to use DialogComponent (used for svg export, pileup sort, etc) on embedded components (@cmdcolin)
- #1945 Fix hic not being able to render due to incorrect lazy loading (@cmdcolin)
- Other
📝 Documentation
- #2002 Add @jbrowse/img to homepage (@cmdcolin)
- #2007 Update docs for modifications/methylation coloring, plugin store, and the sequence panel in feature details (@cmdcolin)
- #1976 reorganize the demo page to emphasize the cancer sv demo more (@rbuels)
- #1952 Add demo for 1000 genomes extended trio dataset to website (@cmdcolin)
- #1862 Add example for using a build-time included plugin to storybook (@cmdcolin)
🏠 Internal
- Other
core,development-tools
Committers: 4
- Colin Diesh (@cmdcolin)
- Elliot Hershberg (@elliothershberg)
- Garrett Stevens (@garrettjstevens)
- Robert Buels (@rbuels)
Release v1.2.0
We're excited to announce the v1.2.0 release of JBrowse Web!
New plugin store added to website
One of the core aspects of JBrowse 2 is that it is an extensible
platform for biological visualization that can be extended with
plugins. We are excited to introduce the first version of our
plugin store, where we list the current external plugins that
are available. Check it out here.
In the coming weeks, we will also be bringing this plugin store directly
into the application, allowing plugin installation with the click of a button.
Stay tuned!
SVG export
We're excited to introduce a new feature to JBrowse Web: built-in
SVG export of track visualizations! This feature currently supports
the linear genome view, and will be extended to more views in future
releases.
With the addition of this feature, it is now even easier to create
publication-ready screenshots of JBrowse views.
Virtualized track lists
An important consideration for genomics software is scaling to very large
datasets. We have implemented a virtualization of our hierarchical track
selector, enabling it to support arbitrarily large track lists.
Lazy loading
We have expanded our use of lazy loading, which optimizes app performance and
improves load times.
1.2.0 (2021-05-03)
Packages in this release
| Package | Download |
|---|---|
| @jbrowse/core | https://www.npmjs.com/package/@jbrowse/core |
| @jbrowse/plugin-alignments | https://www.npmjs.com/package/@jbrowse/plugin-alignments |
| @jbrowse/plugin-bed | https://www.npmjs.com/package/@jbrowse/plugin-bed |
| @jbrowse/plugin-breakpoint-split-view | |
| @jbrowse/plugin-circular-view | https://www.npmjs.com/package/@jbrowse/plugin-circular-view |
| @jbrowse/plugin-config | https://www.npmjs.com/package/@jbrowse/plugin-config |
| @jbrowse/plugin-data-management | https://www.npmjs.com/package/@jbrowse/plugin-data-management |
| @jbrowse/plugin-dotplot-view | |
| @jbrowse/plugin-filtering | |
| @jbrowse/plugin-gff3 | https://www.npmjs.com/package/@jbrowse/plugin-gff3 |
| @jbrowse/plugin-hic | |
| @jbrowse/plugin-legacy-jbrowse | |
| @jbrowse/plugin-linear-comparative-view | |
| @jbrowse/plugin-linear-genome-view | https://www.npmjs.com/package/@jbrowse/plugin-linear-genome-view |
| @jbrowse/plugin-lollipop | |
| @jbrowse/plugin-menus | |
| @jbrowse/plugin-protein | |
| @jbrowse/plugin-rdf | |
| @jbrowse/plugin-sequence | https://www.npmjs.com/package/@jbrowse/plugin-sequence |
| @jbrowse/plugin-spreadsheet-view | |
| @jbrowse/plugin-sv-inspector | |
| @jbrowse/plugin-svg | https://www.npmjs.com/package/@jbrowse/plugin-svg |
| @jbrowse/plugin-trackhub-registry | |
| @jbrowse/plugin-variants | https://www.npmjs.com/package/@jbrowse/plugin-variants |
| @jbrowse/plugin-wiggle | https://www.npmjs.com/package/@jbrowse/plugin-wiggle |
| @jbrowse/cli | https://www.npmjs.com/package/@jbrowse/cli |
| @jbrowse/desktop | |
| @jbrowse/react-linear-genome-view | https://www.npmjs.com/package/@jbrowse/react-linear-genome-view |
| @jbrowse/web |
🚀 Enhancement
core- #1125 Export SVG (@cmdcolin)
- #1867 Virtualized tree for tracklist to support having thousands of tracks (@cmdcolin)
- #1660 Allow connections to have multiple assemblies (@garrettjstevens)
- #1864 Add Material UI's DataGrid to re-exports (@garrettjstevens)
- #1875 Make drawer widget titles stay visible when scrolling inside the widget (@cmdcolin)
- #1877 Add ability to copy the text produced by the feature details sequence panel to MS Word/Google Docs and preserve styling (@cmdcolin)
- #1854 Make "About track" dialog available from tracklist and for non-LGV tracks (@cmdcolin)
- #1853 Add mouseovers in feature details that show field descriptions for VCF fields (@cmdcolin)
- Other
- #1892 Create new JB2 plugin store (@elliothershberg)
- #1901 Make using --out for add-assembly create output directory if it does not exist and fix outputting to symlink (@cmdcolin)
- #1850 Add true breakend ALT strings to the feature details panel (@cmdcolin)
- #1878 Add --delete to set-default-session, fix --session (@garrettjstevens)
- #1861 Change Alignments track "Fade mismatches by quality" setting to a separate config param and made it less strict (@cmdcolin)
🐛 Bug Fix
core- #1924 Fix import of BED and navToLocString from spreadsheet views (@cmdcolin)
- #1918 Fix issue with some falsy values being hidden in feature details (@cmdcolin)
- #1911 Fix breakpoint split view visualizations for files that need ref renaming (e.g. chr1 vs 1) (@cmdcolin)
- #1904 Fix issue with synteny polygons displaying slightly offset (@cmdcolin)
- #1884 Fix rIC ponyfill for use on Safari (@cmdcolin)
- Other
- #1912 Fix reloading of local sessions when using React.StrictMode (@cmdcolin)
- #1900 Make clicking away from autocomplete popup on track container work (@cmdcolin)
- #1878 Add --delete to set-default-session, fix --session (@garrettjstevens)
- #1871 Fix crash on dotplot/linear synteny import form and when closing linear synteny track (@cmdcolin)
- #1860 Fix alignments read filter jexl syntax (@cmdcolin)
📝 Documentation
- #1914 Use MDX to add proper image captions in web and pdf documentation (@cmdcolin)
- #1855 Add download page in website header and new super-quick-start guide (@cmdcolin)
🏠 Internal
core- #1932 Update analytics and share API URLs to refer to more stable locations (@peterkxie)
- #1888 More lazy loading of react components to reduce bundle size ([@cmdcolin](https:...
Release v1.1.0
We're pleased to announce a new release of JBrowse Web!
Changed callbacks language from JavaScript to Jexl
To allow users to safely and seamlessly share advanced configurations in sessions, we now use Jexl to express configuration callbacks. Note that this is a breaking change, function()-style callbacks will no longer work.
For details, see the callbacks section of our configuration guide.
Fetch intron and upstream/downstream sequences
We also have several other improvements including the ability to get intron and
upstream/downstream sequence in the feature details
Interactive documentation using Storybook
Another new update is the first release of our interactive Storybook docs for the embeddable React Linear Genome View.
The docs contain live examples of how the LGV component can be used, along with source-code examples.
The site can be found here.
Enhanced navigation to drawer widget stack
We have added a dropdown to enhance navigation between stack of active widgets. The update also adds a minimize button to allow quick access to full screen JBrowse web.
🚀 Enhancement
core- #1846 Improve copy+paste in the data grids for feature details (@cmdcolin)
- #1814 Add ability to get promoter sequence and intron sequence for genes from the feature details panel (@cmdcolin)
- #1816 Remove some animation effects (@cmdcolin)
- #1778 Adds dropdown to show drawer widget stack (@teresam856)
- #1685 Change callbacks language from JavaScript to Jexl (@peterkxie)
- Other
- #1831 Add dialog for launching breakpoint split view from variant feature details (@cmdcolin)
- #1803 Transcript and gene glyphs can now display implied UTRs, active by default (@cmdcolin)
- #1808 Add another heuristic for returning gene features from BigBed (@cmdcolin)
- #1774 Add warning dialog in LGV before returning to import form to prevent accidentally losing the current view (@cmdcolin)
🐛 Bug Fix
core- #1811 Check for existence of window more robustly to allow in SSR or node applications (@elliothershberg)
- #1793 Fix dotplot rendering outside it's allowed bounds (@cmdcolin)
- #1783 Add hic aborting and fix remoteAbort signal propagation (@cmdcolin)
- #1723 A few bugfixes (@garrettjstevens)
- Other
📝 Documentation
- #1824 Add storybook docs page for nextjs usage (@elliothershberg)
- #1770 1469 storybook deploy (@elliothershberg)
- #1807 Update developer guide to cover displays, and highlight working external plugins (@cmdcolin)
- #1779 Collaborative release announcement editing (@rbuels)
- #1791 Add a couple more demos for our live version with MDX (@cmdcolin)
🏠 Internal
- Other
- #1820 Create v1.1.0.md, draft of release announcements (@garrettjstevens)
- #1823 Add note about previewing changelog to CONTRIBUTING.md (@garrettjstevens)
core
Release v1.0.4
This release of JBrowse Web includes a great many small improvements and bug fixes, see the full changelog below.
Some particularly salient improvements include:
Better indications for insertions
The alignments track received a couple updates including "large insertion indicators" for large indels, and also an upside-down count of clipping or insertion events. There is also a triangular indicator plotted when the insertion/clip count exceeds a threshold at that position defaulted to 30% of reads
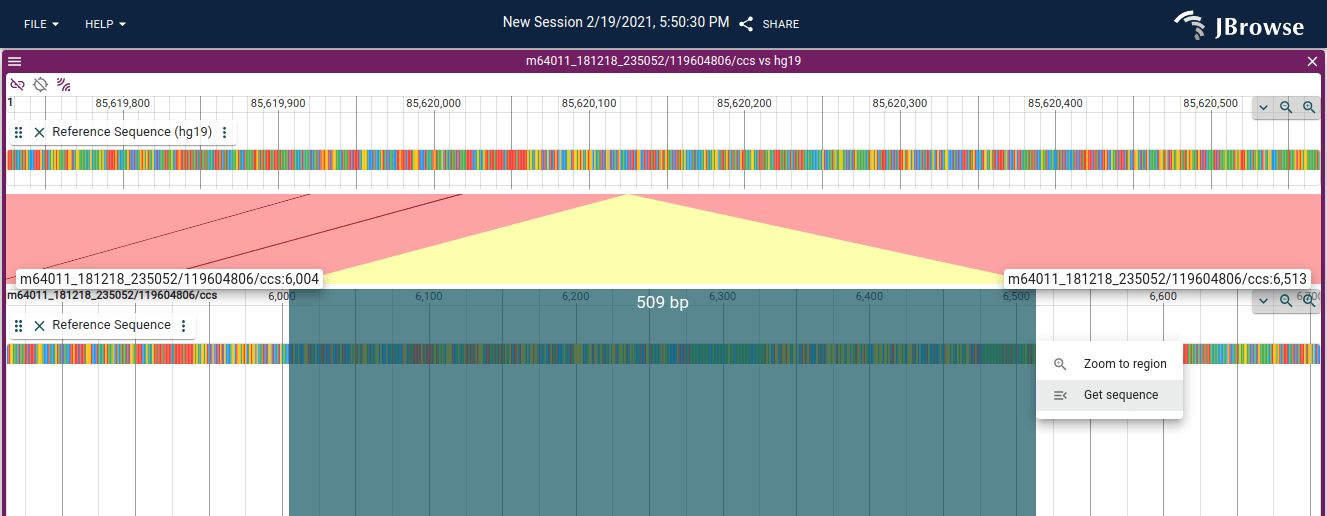
Click and drag the overview bar to "Get sequence"
Users can now download regions of sequence by selecting a region in the linear genome view and clicking "get sequence". See the demonstration video below:
You can also "get sequence" in the read vs reference view, which allows you to "get sequence" for the inserted bases or softclipped bases from a read alignment
A long-requested feature, implemented in #1588 by @teresam856!
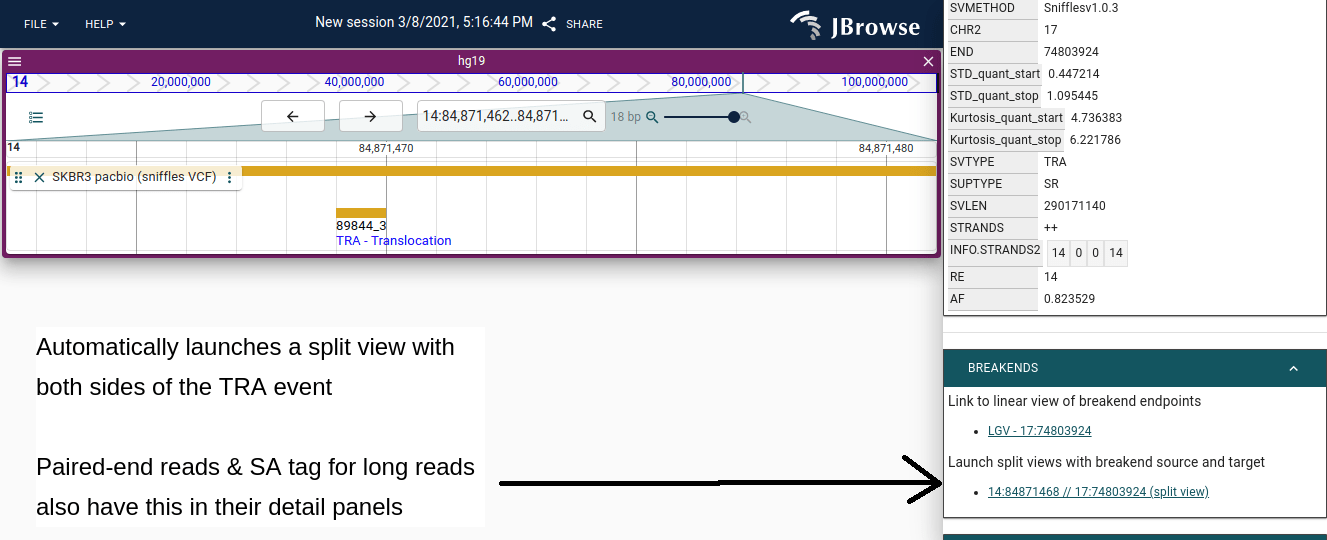
Enhanced navigation of paired end reads and BND/TRA breakends
Feature detail panels for BND/TRA features, long split-alignments, and paired-end reads have links to navigate or popup of the breakpoint split views with their mates.
Implemented by @cmdcolin in #1701.
🚀 Enhancement
core- #1758 Add ability to get stitched together CDS, protein, and cDNA sequences in feature details (@cmdcolin)
- #1721 Manually adjust feature height and spacing on alignments track (@cmdcolin)
- #1728 Add list of loaded plugins to the "About widget" (@rbuels)
- #1711 Add plugin top-level configuration (@teresam856)
- #1699 Add sequence track for both read and reference genome in the "Linear read vs ref" comparison (@cmdcolin)
- #1701 Add clickable navigation links to supplementary alignments/paired ends locations and BND/TRA endpoints in detail widgets (@cmdcolin)
- #1601 Add ability to color by per-base quality in alignment tracks (@cmdcolin)
- #1640 Move stats calculation to BaseFeatureAdapter (@cmdcolin)
- #1588 Add "Get sequence" action to LGV rubber-band (@teresam856)
- Other
- #1743 Add color picker and choice of summary score style for wiggle track (@cmdcolin)
- #1763 Add a "CSS reset" to jbrowse-react-linear-genome-view to prevent parent styles from outside the component leaking in (@cmdcolin)
- #1756 Split alignments track menu items into "Pileup" and "SNPCoverage" submenus (@cmdcolin)
- #1742 Add ability to display crosshatches on the wiggle line/xyplot renderer (@cmdcolin)
- #1736 Fix CLI add-track --load inPlace to put exact contents into the config, add better CLI example docs (@cmdcolin)
- #1394 Add new menu items for show/hide feature labels, set max height, and set compact display mode (@cmdcolin)
- #1720 Standardize phred qual scaling between BAM and CRAM and add option to make mismatches render in a lighter color when quality is low (@cmdcolin)
- #1704 Add "Show all regions in assembly" to import form and make import form show entire region when refName selected (@cmdcolin)
- #1687 Threshold for indicators on SNPCoverage + inverted bargraph of interbase counts for sub-threshold events (@cmdcolin)
- #1695 Improve zoomed-out display of quantitative displays tracks when bicolor pivot is active (@cmdcolin)
- #1680 Add on click functionality to quantitative track features (@teresam856)
- #1630 Get column names from BED tabix files and other utils for external jbrowse-plugin-gwas support (@cmdcolin)
- #1709 Improve sorting and filtering in variant detail widget (@cmdcolin)
- #1688 Bold insertion indicator for large insertions on pileup track (@cmdcolin)
- #1669 Allow plain json encoding of the session in the URL (@cmdcolin)
- #1642 Enable locstring navigation from LGV import form (@teresam856)
- #1655 Add GFF3Tabix and BEDTabix inference to JB1 connection (@garrettjstevens)
- #1643 Add an offset that allows all wiggle y-scalebar labels to be visible (@cmdcolin)
- #1632 Displays warnings when receiving a session with custom callbacks (@peterkxie)
- #1615 Increase pileup maxHeight (@cmdcolin)
- #1624 GCContent adapter (@cmdcolin)
- #1614 Add insertion and clip indicators to SNPCoverage views (part of Alignments tracks) (@cmdcolin)
- #1610 Display error message from dynamodb session sharing error (@cmdcolin)
🐛 Bug Fix
- Other
- #1777 Quick fix for block error (@cmdcolin)
- #1748 External plugins load after confirming config warning (@peterkxie)
- #1750 Fix pileup sorting when using string tag (@cmdcolin)
- #1747 Fix the position of the popup menu after rubberband select when there is a margin on the component e.g. in embedded (@cmdcolin)
- #1736 Fix CLI add-track --load inPlace to put exact contents into the config, add better CLI example docs (@cmdcolin)
- #1731 Fix alignment track ability to remember the height of the SNPCoverage subtrack on refresh (@cmdcolin)
- #1719 Fix for navigation past end of chromosome (@cmdcolin)
- #1698 Fix rendering read vs ref comparisons with CIGAR strings that use = sign matches (@cmdcolin)
- #1697 Fix softclipping configuration setting causing bases to be missed (@cmdcolin)
- #1689 Disable copy/delete menu items for reference sequence track (@teresam856)
- #1682 Fix parsing of BED and BEDPE files with comment header for spreadsheet view (@cmdcolin)
- #1679 Fix issue with using launching the add track widget on views that are not displaying any regions ([@t...
Release v1.0.3
This release of JBrowse Web includes several improvements and bug fixes, including:
- Search and dropdown combined into single intuitive component on linear genome view
- Big alignment track enhancements
- Added sort by tag to sorting options
- Added ability to color by preset options such as "base pair", "mapping quality", and "tag"
- Added ability to filter by preset options such as "read paired" and "read reverse strand"
- Feature detail widget now shows subfeature details
- Added --branch and --nightly flags for create and upgrade CLI commands
- Improved admin server GUI
- Additional track menu options for wiggle and SNP coverage tracks such as "autoscale", "log scale" and "histogram fill"
- Improved build system for external plugins (check out our new plugin template at https://github.com/GMOD/jbrowse-plugin-template)
1.0.3 (2021-01-11)
Packages in this release
🚀 Enhancement
- Other
- #1560 Provide a dialog to add extra genomic context for linear read vs. ref visualization (@elliothershberg)
- #1604 Add ability to filter for read name to the alignments filter dialog (@cmdcolin)
- #1599 Replace 'show all regions' with 'show all regions in assembly' (@cmdcolin)
- #1595 Admin server GUI enhancements (@elliothershberg)
- #1584 Restructure demo page and release cancer demo (@elliothershberg)
- #1579 Create --branch and --nightly flags for
jbrowse createandjbrowse upgradecommands (@cmdcolin) - #1575 Improve mobx-state-tree type validation errors (@cmdcolin)
- #1574 Make softclip indicator black if no seq available (@cmdcolin)
- #1554 Coloring options with simple color for tag (@peterkxie)
- #1565 Rename jbrowse cli add-track --type to --trackType (@cmdcolin)
- #1558 Add docs for sequence track, variant track, launching synteny from dotplot, and add UCSC plugin to demo (@cmdcolin)
- #1533 Display file headers in pre tag in about dialogs and bump @gmod/bam and @gmod/tabix package versions (@cmdcolin)
- #1541 Add more info about adding a PAF file to the synteny import form (@cmdcolin)
- #1509 Combine Search and Dropdown component on LGV (@teresam856)
- #1530 Add spreadsheet filter support for derived columns (@elliothershberg)
- #1483 Add session export to and import from file (@garrettjstevens)
- #1519 Add autoSql to the bigBed "About this track" dialog (@cmdcolin)
core- #1531 Add track menu options for autoscale, log scale, histogram fill, setting min/max score, and zoom level/resolution for wiggle/snpcoverage tracks (@cmdcolin)
- #1473 Color, filter, and sort options for the alignments (@cmdcolin)
- #1576 Add location string to tooltip for wiggle and SNPCoverage tracks (@cmdcolin)
- #1529 Display subfeatures in feature details widget (@cmdcolin)
core,development-tools- #1578 Update build system for external plugins (@garrettjstevens)
🐛 Bug Fix
- Other
- #1608 Take into account offsetX of the rubberband on scalebar zooming (@cmdcolin)
- #1597 Fix crash when there are undefined references in the state tree e.g. when a track is deleted but still referred to by a session (@peterkxie)
- #1598 Disable 'copy to clipboard' while share url being generated (@peterkxie)
- #1589 Fix the display of trackhub registry results (@cmdcolin)
- #1573 Update hic-straw to fix error for hic files with many scaffolds (@cmdcolin)
- #1563 Remove softclip and hardclip from being counted as SNPs in the SNPCoverage (@cmdcolin)
- #1559 Avoid errors from breakpoint split view related to getBoundingClientRect on null track (@cmdcolin)
- #1540 Fix memory leak when side scrolling LGV blocks (@cmdcolin)
- #1534 Fix breakpoint split view showing too many connections for paired end ends (@cmdcolin)
- #1524 Move loading flag for spreadsheet import wizard to volatile to avoid it persisting across refresh (@cmdcolin)
- #1521 Add missing dep to react-linear-genome-view (@garrettjstevens)
core
📝 Documentation
- #1594 Add GFF3 example to quickstart (@cmdcolin)
- #1581 Add some features that are missing from jbrowse 2 to the feature comparison table (@cmdcolin)
- #1558 Add docs for sequence track, variant track, launching synteny from dotplot, and add UCSC plugin to demo (@cmdcolin)
- #1537 Add CONTRIBUTING.md with tips for getting started with codebase (@cmdcolin)
🏠 Internal
Release v1.0.2
This release addresses two main issues:
- A bug introduced in v1.0.1 where the CLI command
jbrowse createwould not
download the latest version of JBrowse Web. - A fix for a problem that prevented
@jbrowse/react-linear-genome-view(first
released in v1.0.1) from working properly.
Several other bug fixes and improvements are also included. See details below.
v1.0.2 (2020-12-02)
Packages in this release
| Package | Download |
|---|---|
| @jbrowse/core | https://www.npmjs.com/package/@jbrowse/core |
| @jbrowse/development-tools | https://www.npmjs.com/package/@jbrowse/development-tools |
| @jbrowse/plugin-alignments | https://www.npmjs.com/package/@jbrowse/plugin-alignments |
| @jbrowse/plugin-bed | https://www.npmjs.com/package/@jbrowse/plugin-bed |
| @jbrowse/plugin-breakpoint-split-view | |
| @jbrowse/plugin-circular-view | https://www.npmjs.com/package/@jbrowse/plugin-circular-view |
| @jbrowse/plugin-config | https://www.npmjs.com/package/@jbrowse/plugin-config |
| @jbrowse/plugin-data-management | https://www.npmjs.com/package/@jbrowse/plugin-data-management |
| @jbrowse/plugin-dotplot-view | |
| @jbrowse/plugin-filtering | |
| @jbrowse/plugin-gff3 | https://www.npmjs.com/package/@jbrowse/plugin-gff3 |
| @jbrowse/plugin-hic | |
| @jbrowse/plugin-legacy-jbrowse | |
| @jbrowse/plugin-linear-comparative-view | |
| @jbrowse/plugin-linear-genome-view | https://www.npmjs.com/package/@jbrowse/plugin-linear-genome-view |
| @jbrowse/plugin-lollipop | |
| @jbrowse/plugin-menus | |
| @jbrowse/plugin-protein | |
| @jbrowse/plugin-rdf | |
| @jbrowse/plugin-sequence | https://www.npmjs.com/package/@jbrowse/plugin-sequence |
| @jbrowse/plugin-spreadsheet-view | |
| @jbrowse/plugin-sv-inspector | |
| @jbrowse/plugin-svg | https://www.npmjs.com/package/@jbrowse/plugin-svg |
| @jbrowse/plugin-trackhub-registry | |
| @jbrowse/plugin-wiggle | https://www.npmjs.com/package/@jbrowse/plugin-wiggle |
| @jbrowse/cli | https://www.npmjs.com/package/@jbrowse/cli |
| @jbrowse/desktop | |
| @jbrowse/protein-widget | |
| @jbrowse/react-linear-genome-view | https://www.npmjs.com/package/@jbrowse/react-linear-genome-view |
| @jbrowse/web |
🚀 Enhancement
core- #1513 Add a custom scrollbar that overrides the auto-hiding behavior of scrollbars on OSX (@elliothershberg)
🐛 Bug Fix
- Other
- #1514 react-linear-genome-view bug fixes (@garrettjstevens)
- #1517 Fix the use of filtering display on desktop (@cmdcolin)
- #1512 Fix setting maxDisplayedBpPerPx for pileup display, helps prevent too large an area from being rendered (@cmdcolin)
- #1442 Change track selector togglebutton to normal button (@cmdcolin)
- #1506 Fix horizontally flipped translation frames position (@cmdcolin)
- #1501 Fix CLI to allow jbrowse create to download newer monorepo tag format (@cmdcolin)
core
Committers: 3
- Colin Diesh (@cmdcolin)
- Elliot Hershberg (@elliothershberg)
- Garrett Stevens (@garrettjstevens)
Release v1.0.1
This release of JBrowse Web includes several improvements and bug fixes,
including:
- A new sequence track renderer that includes translation frames
- The addition of session connections
- A new track selector icon
- Improvements to feature details widgets
- And more! See details below.
To install JBrowse 2 for the web, you can download the release ZIP, or you can
use the JBrowse CLI to automatically download the latest version. See the
JBrowse web quick start for more
details.
See "Packages in this release" in the changelog for links to packages published
on NPM.
v1.0.1 (2020-11-25)
Packages in this release
| Package | Download |
|---|---|
| @jbrowse/core | https://www.npmjs.com/package/@jbrowse/core |
| @jbrowse/development-tools | https://www.npmjs.com/package/@jbrowse/development-tools |
| @jbrowse/plugin-alignments | https://www.npmjs.com/package/@jbrowse/plugin-alignments |
| @jbrowse/plugin-bed | https://www.npmjs.com/package/@jbrowse/plugin-bed |
| @jbrowse/plugin-breakpoint-split-view | |
| @jbrowse/plugin-circular-view | |
| @jbrowse/plugin-config | https://www.npmjs.com/package/@jbrowse/plugin-config |
| @jbrowse/plugin-data-management | https://www.npmjs.com/package/@jbrowse/plugin-data-management |
| @jbrowse/plugin-dotplot-view | |
| @jbrowse/plugin-filtering | |
| @jbrowse/plugin-gff3 | https://www.npmjs.com/package/@jbrowse/plugin-gff3 |
| @jbrowse/plugin-hic | |
| @jbrowse/plugin-legacy-jbrowse | |
| @jbrowse/plugin-linear-comparative-view | |
| @jbrowse/plugin-linear-genome-view | https://www.npmjs.com/package/@jbrowse/plugin-linear-genome-view |
| @jbrowse/plugin-lollipop | |
| @jbrowse/plugin-menus | |
| @jbrowse/plugin-protein | |
| @jbrowse/plugin-rdf | |
| @jbrowse/plugin-sequence | https://www.npmjs.com/package/@jbrowse/plugin-sequence |
| @jbrowse/plugin-spreadsheet-view | |
| @jbrowse/plugin-sv-inspector | |
| @jbrowse/plugin-svg | https://www.npmjs.com/package/@jbrowse/plugin-svg |
| @jbrowse/plugin-trackhub-registry | |
| @jbrowse/plugin-variants | https://www.npmjs.com/package/@jbrowse/plugin-variants |
| @jbrowse/plugin-wiggle | https://www.npmjs.com/package/@jbrowse/plugin-wiggle |
| @jbrowse/cli | https://www.npmjs.com/package/@jbrowse/cli |
| @jbrowse/desktop | |
| @jbrowse/protein-widget | |
| @jbrowse/react-linear-genome-view | https://www.npmjs.com/package/@jbrowse/react-linear-genome-view |
| @jbrowse/web |
🚀 Enhancement
- Other
- #1462 Allow importing gzip and bgzip files in the spreadsheet and SV inspector (@cmdcolin)
- #1460 Add support for more bigBed subtypes and fallback for unsupported types (@peterkxie)
- #1455 Add the ability to use connection across refreshes in jbrowse-web using session connections (@peterkxie)
- #1439 1381 improve assembly add form (@elliothershberg)
- #1433 Make add track warning a bit more lenient (@garrettjstevens)
- #1420 Add the assembly manager feature to jbrowse-desktop (@elliothershberg)
core- #1458 Add three frame translation to the sequence track (@cmdcolin)
- #1453 Change "Factory reset" to "Reset session" in jbrowse-web (@teresam856)
- #1441 New icon for the track selector (@cmdcolin)
- #1438 Improve assembly loading time by moving to main thread (@cmdcolin)
- #1434 Create separate config schema for ReferenceSequenceTrack (@elliothershberg)
🐛 Bug Fix
core- Other
- #1489 Fix long read vs ref CIGAR rendering for horizontally flipped synteny view (@cmdcolin)
- #1460 Add support for more bigBed subtypes and fallback for unsupported types (@peterkxie)
- #1472 Wait on assemblies that are being tracked by the assemblyManager only (@cmdcolin)
- #1466 Avoid rendering the display and renderer settings in the about this track dialog (@cmdcolin)
- #1461 Fix usage of jbrowse-cli on node 10.9 related to fs.promises (@cmdcolin)
- #1452 Bug: search box disappears from LGV header on smaller widths (@teresam856)
- #1432 Make global variables window.JBrowseSession and window.JBrowseRootModel available in jbrowse-web (@teresam856)
- #1431 Fix connection tracks not showing up in track selector (@garrettjstevens)
- #1428 Fix the listVersions behavior of the jbrowse-cli returning duplicate entries (@cmdcolin)
- #1422 Fix crash from empty ALT field in VCF (@cmdcolin)
- #1413 Fix ability to add CRAM tracks using the web based add-track GUI (@cmdcolin)
📝 Documentation
- #1435 Updates to quickstart guides (@garrettjstevens)
🏠 Internal
- Other
- #1437 Use lerna-changelog for changelog generation (@cmdcolin)
- #1465 Establish minimum node version of 10.4 for using jbrowse-cli tools (@cmdcolin)
- #1454 Fix GH workflow build (@elliothershberg)
- #1448 Move building and testing from Travis to GitHub Workflow (@garrettjstevens)
- #1450 Fix website build (@garrettjstevens)
core- #1468 Have assembly manager get plugin manager from factory function args (@garrettjstevens)
Committers: 5
- Col...