Ben Bodek, Brian Chen, Forrest Hurley, Brian Richardson, Emmanuel Rockwell
The HOOVS package (“High-Dimensional Ordinal Outcome Variable
Selection”) is for a group project for Bios 735 (statistical computing).
The goal of the project is to develop two methods to perform variable
selection on a high-dimensional data set with an ordinal outcome. The
first method is a LASSO-penalized ordinal regression model and the
second is a random forest model.
Installation of the HOOVS from GitHub requires the
devtools
package and can be done in the following way.
# Install the package
devtools::install_github("brian-d-richardson/HOOVS")# Then load it
library(HOOVS)Other packages used in this README can be loaded in the following chunk.
suppressPackageStartupMessages(if (!require(dplyr)) {install.packages("dplyr")})
suppressPackageStartupMessages(if (!require(tidyr)) {install.packages("tidyr")})
suppressPackageStartupMessages(if (!require(ggplot2)) {install.packages("ggplot2")})
suppressPackageStartupMessages(if (!require(ordinalNet)) {install.packages("ordinalNet")})
suppressPackageStartupMessages(if (!require(foreign)) {install.packages("foreign")})
suppressPackageStartupMessages(if (!require(devtools)) {install.packages("devtools")})
suppressPackageStartupMessages(if (!require(tictoc)) {install.packages("tictoc")})
suppressPackageStartupMessages(if (!require(psych)) {install.packages("psych")})
load_all()
#> ℹ Loading HOOVS
#For reproducibility
set.seed(1)To build the final report document render final_presentation.Rmd.
We begin with a theoretical introduction of the ordinal regression model
and how the HOOVS package calculates parameter estimates.
Suppose that, for observation , the ordinal outcome
given the covariate vector
has a multinomial distribution with
outcome categories and probabilities of success
. That is,
The cumulative probability for subject
and ordinal outcome category
is
. Note
that by definition
.
The following proportional odds model relates the cumulative probability
for subject
and ordinal outcome category
to the covariates
via the parameters
and
with a logit link
function.
In this model, are outcome category-specific
intercepts for the first
ordinal outcome categories and
are the slopes corresponding to the
covariates. Since the cumulative probabilities must be increasing
in
, i.e.,
, we
require that
.
The likelihood function for the ordinal regression model is
Let
be the standardized log-likelihood.
The LASSO-penalized ordinal regression model is fit by minimizing the
following objective function with respect to
and
.
The objective function can be minimized using a proximal gradient descent (PGD) algorithm.
Fix the following initial parameters for the PGD algorithm: (the initial step size),
(the step size decrement value), and
(the convergence criterion).
The proximal projection operator for the LASSO penalty (applied to
but not to
) is
Given current estimates ,
search for updated estimates
by following the steps:
Continue updating
until convergence, i.e., until
.
A technical note is that the
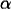 parameters are constrained by
parameters are constrained by 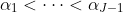 . We can reparametrize the model
with
. We can reparametrize the model
with 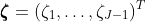 , where 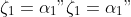 and for
. Then
have no constraints. So we can
follow the above procedure to minimize the above objective function with
respect to
and
, then back-transform to obtain estimates for
.
The HOOVS package allows the user to simulate their own data with the
simulate.data() function. For
subjects, we generate
covariates from independent standard normal distributions[1].
Given true parameters
and
, we compute the multinomial probabilities for the
outcome for each individual and simulate
accordingly.
# sample size
n <- 1000
# number of covariates
p <- 50
# number of categories for ordinal outcome
J <- 4
# grid of lambdas
lambdas <- seq(0.2, 0, -0.02)
# set population parameters
alpha <- seq(.5, 4, length = J - 1) # category-specific intercepts
beta <- rep(0, p) # slope parameters
beta[1: floor(p / 2)] <- 1 # half of the betas are 0, other half are 1
# simulate data according to the above parameters
dat <- simulate.data(
n = 1000,
alpha = alpha,
beta = beta)For this example, we simulated data with
= 1000 observations,
= 50 covariates,
= 4 ordinal outcome categories, and true parameter values of
= (0.5, 2.25, 4) and
= (1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0). Note that this implies the first half of the
covariates are truly associated with the outcome and the last half are
not. The first 10 rows and 10 columns of the data set are shown below.
dat[1:10, 1:10] %>%
mutate_if(.predicate = function(x) is.numeric(x),
.funs = function(x) round(x, digits = 2))
#> y X1 X2 X3 X4 X5 X6 X7 X8 X9
#> 1 3 -0.63 1.13 -0.89 0.74 -1.13 -1.52 -0.62 -1.33 0.26
#> 2 3 0.18 1.11 -1.92 0.39 0.76 0.63 -1.11 0.95 -0.83
#> 3 3 -0.84 -0.87 1.62 1.30 0.57 -1.68 -2.17 0.86 -1.46
#> 4 2 1.60 0.21 0.52 -0.80 -1.35 1.18 -0.03 1.06 1.68
#> 5 4 0.33 0.07 -0.06 -1.60 -2.03 1.12 -0.26 -0.35 -1.54
#> 6 1 -0.82 -1.66 0.70 0.93 0.59 -1.24 0.53 -0.13 -0.19
#> 7 3 0.49 0.81 0.05 1.81 -1.41 -1.23 -0.56 0.76 1.02
#> 8 1 0.74 -1.91 -1.31 -0.06 1.61 0.60 1.61 -0.49 0.55
#> 9 1 0.58 -1.25 -2.12 1.89 1.84 0.30 0.56 1.11 0.76
#> 10 1 -0.31 1.00 -0.21 1.58 1.37 -0.11 0.19 1.46 -0.42Now run our version of a LASSO-penalized ordinal regression function on
the simulated data for various values of
: 0.2, 0.18, 0.16, 0.14, 0.12, 0.1, 0.08, 0.06, 0.04, 0.02,
0.
# test HOOVS LASSO-penalized ordinal regression function
tic("HOOVS ordreg.lasso() function")
res.ordreg <- ordreg.lasso(
formula = y ~ .,
data = dat,
lambdas = lambdas
)
toc()
#> HOOVS ordreg.lasso() function: 5.943 sec elapsed
coef.ordreg <- cbind(res.ordreg$alpha, res.ordreg$beta)We can now look at how the parameter estimates from our funciton change
as the penalty parameter
changes
Note in the above plot that the
estimates do not shrink all the way to 0 since they
are not penalized in the LASSO model. On the other hand, the
estimates do shrink to 0 as
increases. Recall that the data were simulated according to
a model where half of the
values are truly 0 and the other half are truly 1. It
is clear in the above plot which covariates are truly not associated
with the outcome based on how fast their corresponding parameter
estimates shrink to 0.
We can assess the predictive performance of the model with a weighted
kappa statistic. The following table gives the weighted kappa values for
the models fit using each supplied penalty parameter
.
data.frame("lambda" = lambdas,
"Weighted Kappa" = res.ordreg$kappa)
#> lambda Weighted.Kappa
#> lambda=0.2 0.20 0.00000000
#> lambda=0.18 0.18 0.00000000
#> lambda=0.16 0.16 0.00000000
#> lambda=0.14 0.14 0.00000000
#> lambda=0.12 0.12 0.00000000
#> lambda=0.1 0.10 0.00000000
#> lambda=0.08 0.08 0.02319807
#> lambda=0.06 0.06 0.44565785
#> lambda=0.04 0.04 0.72450154
#> lambda=0.02 0.02 0.83576007
#> lambda=0 0.00 0.90220833Suppose we have identified a subset of relevant predictors and want to
perform inference or hypothesis testing using an independent test data
set. We obtain the asymptotic covariance of the parameter estimates from
an ordinal regression model fit with no LASSO penalty by specifying
return.cov = TRUE. Then the standard errors of the parameter estimates
are the square roots of the diagonal entries of the covariance matrix.
# simulate test data
dat <- simulate.data(
n = 500,
alpha = alpha,
beta = beta)
# specify relevant parameters
# (in practice these would be selected using training data)
rel.betas.ind <- which(beta != 0)
# fit model with no penalty
res.ordreg.test <- ordreg.lasso(
formula = y ~ .,
data = select(dat, c("y", paste0("X", rel.betas.ind))),
lambdas = 0,
return.cov = T
)
# covariance matrix
#res.ordreg.test$cov
# standard error of parameter estimates
sqrt(diag(res.ordreg.test$cov))
#> alpha1 alpha2 alpha3 X1 X2 X3 X4 X5
#> 0.1885505 0.2302522 0.3188871 0.1518158 0.1493560 0.1437412 0.1492866 0.1425663
#> X6 X7 X8 X9 X10 X11 X12 X13
#> 0.1532874 0.1598190 0.1501358 0.1533953 0.1471709 0.1463651 0.1503864 0.1473245
#> X14 X15 X16 X17 X18 X19 X20 X21
#> 0.1385858 0.1596688 0.1555481 0.1417651 0.1514717 0.1404234 0.1415255 0.1427013
#> X22 X23 X24 X25
#> 0.1532489 0.1518607 0.1467264 0.1558101- the current version can only handle continuous covariates, and categorical variables must be created into dummy variables by hand.
