-
Notifications
You must be signed in to change notification settings - Fork 0
EVE Project Background
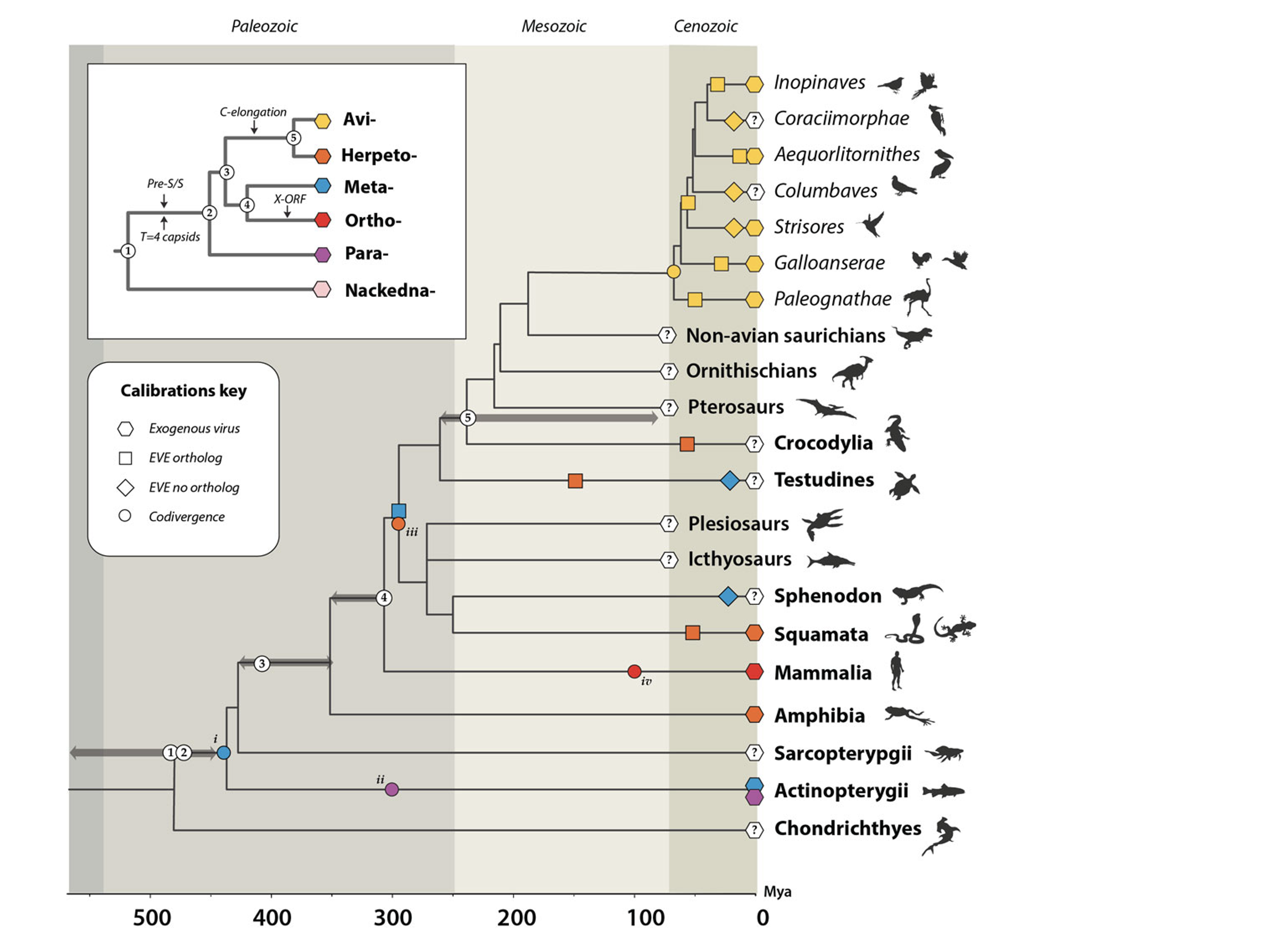
Macroevolutionary timeline of hepadnavirus evolution. The inset panel shows the evolutionary relationships of hepadnavirus genera, the larger tree is a time-calibrated evolutionary tree of vertebrates. Geological eras are indicated by background shading. The scale bar shows time in millions of years before present. For details see Lytras et al (2020). Mya=Million years ago.
Endogenous viral elements (EVEs) are virus-derived sequences that occur in the germline genomes of cellular organisms. EVEs derived from a diverse range of virus families have been identified, and in vertebrate species, these include hepadnaviruses. These ‘endogenous hepatitis B viruses’ (eHBVs) are thought to have originated via ‘germline incorporation’ events in which DNA sequences derived from ancient hepadnaviruses were integrated into chromosomal DNA of germline cells and subsequently inherited as novel host alleles. eHBV sequences provide a source of retrospective information about the distant ancestors of modern hepadnaviruses.
Progress in characterising eHBVs has been hampered by the challenges encountered attempting to analyse their fragmentary and degenerated sequences. Hepadnavirus-GLUE-EVE aims to address these issues.
-
Evolution: EVEs provide insights into the long-term evolutionary history of virus groups, which greatly influences how we understand their biology.
-
Genomics: Some eHBVs may have, or have had, functional roles as host alleles. The prevalence of multicopy eHBV lineages in some species suggests that germline incorporation of hepadnavirus sequences might have significantly influenced the evolution of host genomes.
-
Virus discovery: When new hepadnavirus species are identified, inclusion of EVEs in phylogenetic analyses can provide useful contextual information. Addionally, the eHBV sequences collated here can be used to exclude any potential 'false positive' hits that arise in metagenomics-driven virus discovery efforts (i.e. sequences that seem to represent new hepadnaviruses but in fact derive from genomic DNA).
-
EVE discovery: Using the eHBV sequences provided here as 'probes' for in silico genome screening may enable the detection of novel eHBV elements.
The Hepadnavirus-GLUE-EVE project is designed to provide a comprehensive and accessible resource for the analysis of eHBV sequences. Key features include:
-
GLUE Framework Integration: As part of the Hepadnavirus-GLUE project, the information contained in this repository can be accessed via GLUE's command layer, minimizing the need for labor-intensive pre-processing and enabling streamlined workflows.
-
Standardized Nomenclature: A consistent naming system for eHBV loci to ensure clarity and interoperability across studies.
-
Genomic Mapping: A detailed map showing the precise genomic locations of all known eHBV loci in published whole genome sequence assemblies, aiding in comparative genomic studies.
-
Paleovirus Consensus Genomes: Annotated consensus genomes for the ancient hepadnaviruses that gave rise to the eHBVs, offering a reconstructed view of these viral ancestors.
-
Sequence Alignments: Pre-aligned eHBV sequences and modern hepadnavirus genomes to facilitate comparative analysis.
-
Evolutionary Phylogenies: Phylogenetic trees illustrating the evolutionary relationships between eHBV sequences and extant hepadnaviruses, shedding light on the long-term co-evolution of these viruses with their hosts.