-
Notifications
You must be signed in to change notification settings - Fork 5
CTFileViewer
To access the CT text file viewer, select a sequence folder on the left-hand-side main window pane, and then click on any of the buttons labeled (3) in the screenshot below. Note that the integrated CT file viewer in RNAStructViz will display the sequence and structure pairing data in CT style format regardless of the input file format type used to load the structure.
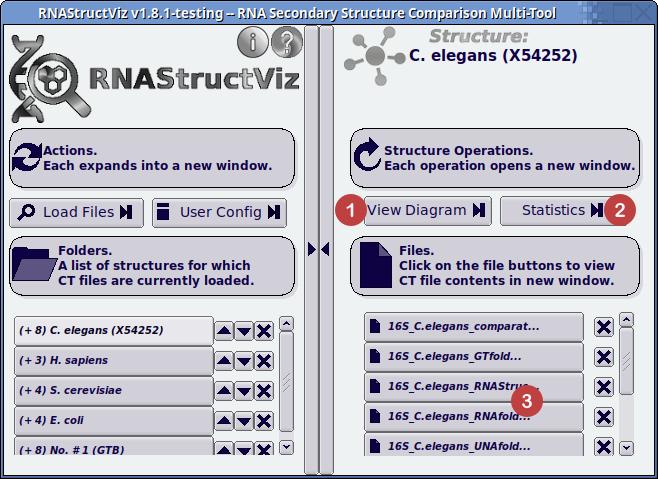
- Click button to view the Diagram Window.
- Click button to open the Statistics Window.
- Click on the particular structure in the selected folder to open the CT-formatted file viewer (topic of this page).
When you click on any of the buttons listed under the Files label, a color-coded plaintext view of the relevant CT file will appear in a separate window. An example depicting the file 16S_C.elegans_GTfold.ct (included by default as a sample structure distributed with RNAStructViz) is shown below:

RNAStructViz has been developed by the Georgia Tech Research Group in Discrete Mathematics and Molecular Biology (gtDMMB) directed by Professor Christine Heitsch (current credits and citations). All communication about running our software, including instantiating bug reports, feature requests, wiki edits, and general inquiries, is logged via our GitHub issues page. Please view the detailed instructions before posting a new issue about support requests.
Source Code Update Information (JSON Format): 📑 Most Recent Commit Activity | 🔖 Latest Release Tag | 📋 Lines of Code