-
Notifications
You must be signed in to change notification settings - Fork 0
Core Schema
In GLUE, information about nucleotide sequence data is structured using a fixed set of object types and relationships known as the Core Schema. This schema organizes virus sequence data in a way that highlights encoded information and evolutionary connections, making it easier to analyze and interpret genomic data. The main elements of the schema are objects, fields, and relationships, which together form the foundation for building and managing a dataset.
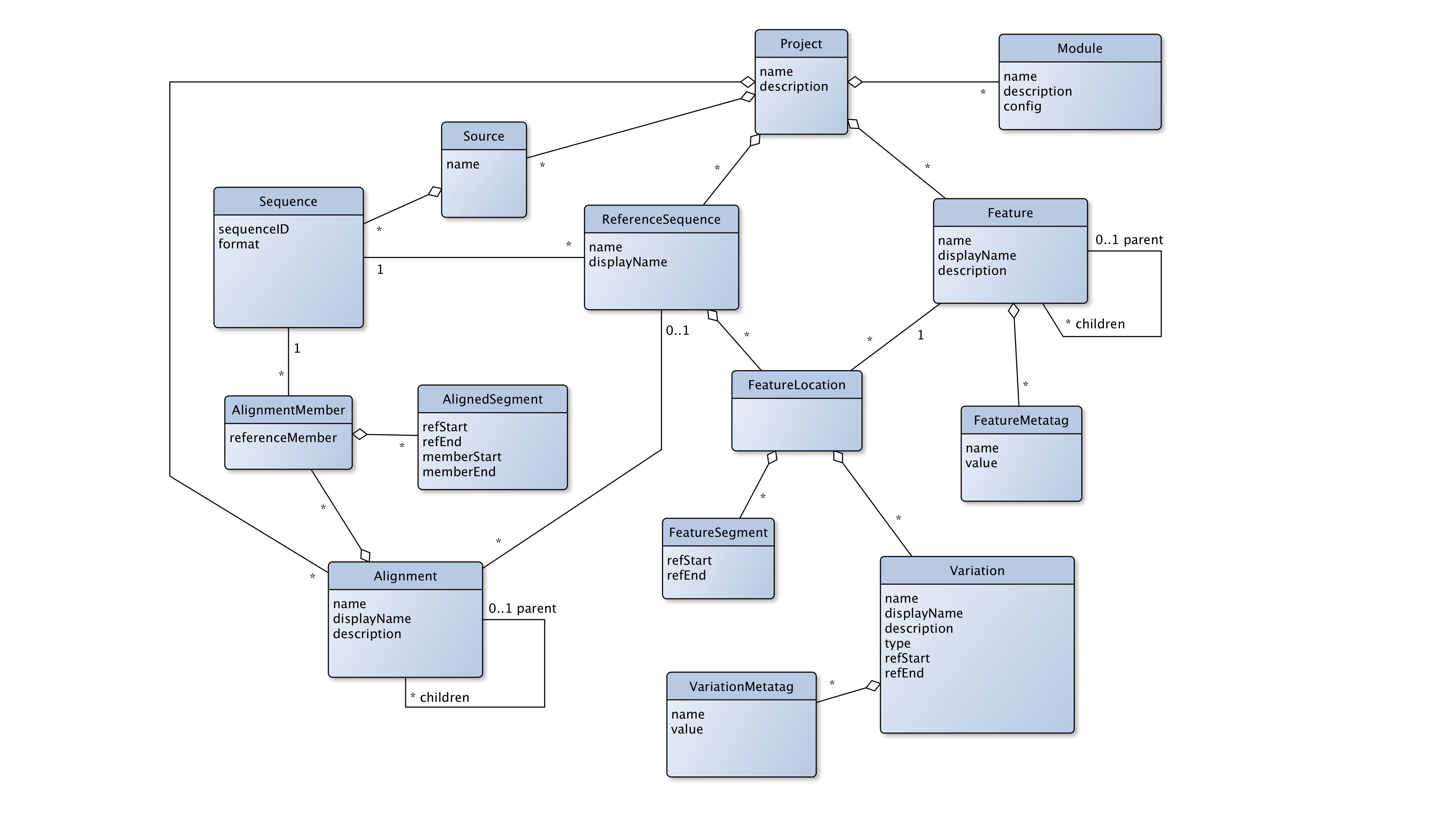
The GLUE core schema. The main object types, fields and relationships in the core schema of GLUE, represented as a Unified Modelling Language (UML) entity-relationship diagram. Object types are represented as blue boxes with fields specified inside. Relationships between types are represented as lines connecting the associated object types. A diamond indicates a composition relationship. Relationship lines are annotated with “multiplicities” indicating how many objects participate in a single instance of the relationship; an asterisk indicates any number of objects
GLUE stores information in the form of objects. Each object can contain fields (specific pieces of data) and relationships (connections to other objects). These objects are defined in the schema and can be extended in specific projects. Below are the key object types:
-
Sequences and Sources: Sequences represent viral nucleotide data and form the basis of each GLUE project. These sequences can come from lab data, computational reconstructions, or consensus sequences. Each sequence is grouped under a Source, which is simply a container to organize sequences.
-
Features: Genomic regions with biological significance, like genes or non-coding regions, are defined as Features. These can be arranged hierarchically, such as coding regions within a polyprotein. Features may have associated metadata (called FeatureMetatags) that helps GLUE perform specialized tasks, like protein translation.
-
ReferenceSequences and FeatureLocations: ReferenceSequences act as anchor points for organizing and interpreting other sequences (usually via an alignment). They link to FeatureLocations, which represent where a genomic region (Feature) is located within a reference sequence. These locations may be made up of multiple segments, allowing for the precise mapping of regions, even those that are discontiguous.
-
Alignments and AlignmentMembers: Sequences in GLUE can be aligned to show nucleotide homologies. An Alignment groups related sequences and uses a reference coordinate space to define how different sequences match up. Each member of an alignment (an AlignmentMember) is mapped to this space, allowing the comparison of homologous regions across sequences.
-
Variations: These objects define specific nucleotide or amino acid patterns. Variations are tied to specific FeatureLocations within reference sequences and allow researchers to quickly analyze viral data for the presence of documented genetic patterns.
Through these object types, GLUE offers a structured way to manage virus sequence data, making it easier for users to perform complex genomic analyses.
 Parvovirus-GLUE by Robert J Gifford Lab.
Parvovirus-GLUE by Robert J Gifford Lab.
For questions, issues, or feedback, please open an issue on the GitHub repository.
- Amdoparvovirus
- Dependoparvovirus
- Erythroparvovirus
- Protoparvovirus
- Copiparvovirus
- Tetraparvovirus
- Bocaparvovirus
- Aveparvovirus
- Chaphamaparvovirus
- Background
- Schema Extensions
- Sequence Data
- Reference Sequences
- Metadata
- Alignments
- Nomenclature
- Phylogenies
- Command Mode Reference
- Mode Non-Specific Commands
- Module Type Reference
- Related Publications
- ICTV Parvoviridae