-
Notifications
You must be signed in to change notification settings - Fork 0
Preprocessing
WARNING: This document is outdated. It will be updated soon.
- TODO: Remove
module use-commands from this document. Refer instead to the setup instructions.
This page is short reference on the module mmpreprocessingpy for expert users. You can find a detailed step-by-step introduction on how to generated the template-config and template-image here.
- Preprocessing registers and splits the full-frame OME-TIFF stack that was recorded with MicroManager into separate TIFF-stacks for each growthlane (GL) in the image. These GLs images are processed using MoMA and are referred to as GL-ROIs (ROI: region of interest).
- The registration aligns the GLs between each frame to compensate for drift that can occur during acquisition. GL-ROIs that sit on the edge of the full-frame image can (partially) move outside of the image due to image drift. A GL-ROI that moves partially outside the full-frame image at any time step of the movie will not be stored.
IMPORTANT: Editing your .bashrc
When using the preprocessing, you have to comment in your .bashrc the
"module use" and "ml MoMA"statements that refer to the MoMA version
that you are using. As an example you would do ('#' comments a line in
bash-scripts):
#module use {PATH_TO_MOMA_MODULES}
#ml MoMATo continue using MoMA after preprocessing, you need to comment the
module use line for the preprocessing and again uncomment line for
using MoMA (i.e. removing the '#' in the example above).
To use the current preprocessing module add this line to your .bashrc :
module use /scicore/home/nimwegen/GROUP/Moma/MM_Analysis/builds/unstable_20230405__FOR_PREPROCESSING_ONLY__mmpreprocesspy_v0.4.0-5b062368/ModulesThis is an example script for launching the preprocessing on Scicore. See below for explanations of the parameters:
#!/bin/bash
DIR="$( cd "$( dirname "${BASH_SOURCE[0]}" )" >/dev/null 2>&1 && pwd )"
SCRIPT_NAME_FULL="$(basename "$0")"
DATE="$(date '+%Y%m%d-%H%M%S')"
SCRIPT_NAME="${SCRIPT_NAME_FULL%.*}"
PREPROC_DIR_TPL="${DIR}/${SCRIPT_NAME}__output__${DATE}/mmpreproc_%s/"
RAW_PATH="/scicore/home/nimwegen/GROUP/MM_Data/Lis/20220128/20220128_VNG1040_AB2h_3/20220128_VNG1040_AB2h_3_MMStack.ome.tif"
FLATFIELD_PATH="/scicore/home/nimwegen/GROUP/MM_Data/Lis/20220128/20220128_gfpflatfield_1/20220128_gfpflatfield_1_MMStack.ome.tif"
POS_NAMES=(Pos0 Pos1)
#TMIN=0
#TMAX=10
GL_DETECTION_TEMPLATE_PATH="$DIR/TEMPLATE/template_config.json"
ROTATION=90.1
IMAGE_REGISTRATION_METHOD=2
FRAMES_TO_IGNORE=(131,189,221,277,338,453,615)
NORMALIZATION_CONFIG_PATH="true"
NORMALIZATION_REGION_OFFSET=120
INTENSITY_NORMALIZATION_RANGE_CUTOFFS=(0,10000)
#FORCED_INTENSITY_NORMALIZATION_RANGE=(1395.39,5859.17)
## DO NOT EDIT BELOW THIS LINE ##
# fixed parameters:
ROI_BOUNDARY_OFFSET_AT_MOTHER_CELL=0
# generate an array of same length as position array, with every element containing the ROTATION scalar value
ROTATIONS=$ROTATION
for f in `seq ${#POS_NAMES[*]}`
do
f=`echo $f - 1 | bc`
ROTATIONS[$f]=$ROTATION
done
module purge
module load MMPreproc
source mm_dispatch_preprocessing.sh
mm_dispatch_preprocessingThe parameters are:
-
PREPROC_DIR_TPL: The output path of the preprocessing script. In this case we set it dynamically relative to the name/location of this script and the data. It can also be fixed. Note that the part/mmpreproc_%s/is ignored (this is for compatibility to the legacy preprocessing). -
RAW_PATH: Path to the OME-TIFF stack containing the measurement. -
FLATFIELD_PATH: Path to the flat-field image corresponding toRAW_PATH. -
POS_NAMES: An array of the position names that should be preprocessed. The names corresponds to the position names in MicroManager. -
TMINandTMAX:-
TMIN: Sets the start frame in the OME-TIFF stack. This index is 0-based, so you will have to subtract 1 from the frame index reported by MicroManager and ImageJ. Commenting this value, sets it to the first frame of the OME-TIFF stack. -
TMAX: Sets the end frame in the OME-TIFF stack. This index is 0-based, so you will have to subtract 1 from the frame index reported by MicroManager and ImageJ. Commenting this value, sets it to the last frame of the OME-TIFF stack. - NOTE: The parameters
TMINandTMAXallow to restrict the range of frames that will be processed by the preprocessing. This allows to e.g. ignore that frames towards the end of an experiment, where the image became defocused. Setting e.g.TMAX=10is also helpful for faster testing of the preprocessing settings (i.e. the registration template, etc.).
-
-
ROTATION[degrees]: The rotation angle of the full-frame image in degrees. See the section "Configuring the GL detection template" on how to determine this value. -
GL_DETECTION_TEMPLATE_PATH: Path to the template that is used for detecting the GLs inside the full-frame. See the section "Configuring the GL detection template". -
NORMALIZATION_CONFIG_PATH: Sets whether to perform normalization. "true": Perform normalization. "false": Do not perform normalization. -
NORMALIZATION_REGION_OFFSET[pixel]: Sets the margin in pixels at both ends of the GL-ROI for the region, where the image normalization will be calculated. -
INTENSITY_NORMALIZATION_RANGE_CUTOFFS: This is a pair/tuple of intensity values (ie.: (min_value, max_value)) with same units as the raw intensities in the input image. When detecting the normalization range, peaks below/above this range will not be considered during detection of the normalization range. This parameter is helpful, when normalizing positions with air-filled objects within the normalization region (e.g. such as number or non-primed GLs). These objects are very bright and break normalization otherwise. This parameter can be removed, when not needed. -
FORCED_INTENSITY_NORMALIZATION_RANGE: This parameter overrides the automatic detection of the normalization range. When used, the normalization range will be set to the specified minimum-/maximum-values for all frames. When used, no normalization logs are stored during preprocessing. This parameter is helpful, when there are not empty GL within the normalization region and hence the image could not be normalized otherwise. It is recommended to not use this parameter unless necessary, because fixing the normalization range (e.g. based on the first frame) can be problematic, when intensities change in later frames (e.g. due to changes in focus or illumination). -
IMAGE_REGISTRATION_METHOD: Sets the method for registering each frame of the TIFF-stack to correct for drift during the acquisition. Valid values are '1' and '2':- '1': Each frame in the TIFF stack is registered to the first frame of the TIFF stack. This is useful when registration fails for a few, isolated frames (e.g. because they are defocused). In this case the remaining frames are still registered correctly. This is the preferred method to use.
- '2': Each frame in the TIFF stack is registered to the previous frame. This can be useful, when images gradually change over the series and that change is strong (e.g. because of a gradual defocus (though not too severe to mess up registration all together) or stage drift, which over time causes a strong positional mismatch to the first frame). In that situation later frames can become so dissimilar to the first frame that registration fails for method 1, but they can still be registered consecutively using this method.
-
FRAMES_TO_IGNORE: This specifies the indices of frames, which will be skipped during preprocessing. The frame index is 1-based like in MicroManager and ImageJ, so that the frame indices correspond to the ones in MicroManager or ImageJ. Skipped frames are replaced by the last non-skipped frame previous to the skipped frame to avoid. Filling in the skipped frames avoids jumps in the time series. IMPORTANT: The specified skipped frames are applied to all positions given inPOS_NAMES. If you have a different sets of skip-frames for different positions you will need to create a separate preprocessing script for each set of positions and their respective skip-frames.
Debugging parameters:
-
ROI_BOUNDARY_OFFSET_AT_MOTHER_CELL: This defines a margin in pixels at the position of the mother cell (i.e. at the closed end of the GL). Increasing this value will grow the ROI in direction of the closed end beyond the position value defined in the template. This value should be kept at 0 unless needed for debugging; defining the ROI size should be done exclusively through the GL detection template (see below).
mmpreprocessingpy can use a template-image to do template-matching in order to detect growthlanes in the mother-machine image. This is used to detect growth lanes in the full frame TIFF-stack. The template matching is done on the first frame of each processed position. For this it uses a template image and configuration file.
Here we briefly describe, how to generate an adequate template image and setup up a corresponding config file.
This is an example config file for template matching which was generate for the experiment 20211117_VNG1040_AB2h_2h_1:
{
"name": "template__20211117_VNG1040_AB2h_2h_1",
"description": "",
"template_image_path": "./TEMPLATE_IMAGE_pos0_frame47__20211117_VNG1040_AB2h_2h_1-1.tif"
"pixel_size_micron": 1.0,
"gl_regions": [
{
"first_gl_position_from_top": 44,
"gl_spacing_vertical": 107.1176,
"horizontal_range": [
55,
660
],
"gl_exit_orientation": "right"
},
{
"first_gl_position_from_top": 41,
"gl_spacing_vertical": 106.9412,
"horizontal_range": [
885,
1460
],
"gl_exit_orientation": "left"
}
],
}This is the corresponding template image as an example of what a template image can look like:

Note that the fact that we have cells in the GLs (usually) does not affect registration performance ("usually" means we have not come across such a case).
Parameter explanation:
-
"name"and"description": These are for documentation purposes for the user (they are not relevant to the functionality and can be left empty). -
"template_image_path": Defines the path to the template image. This path can be relative or absolute. I suggest making this path relative (as shown here) and storing the template image next to the corresponding config-file. This way they can be copied together and used as is. -
"pixel_size_micron": The parameters"horizontal_range","gl_spacing_vertical"and"fist_gl_position_from_top"are expected to be in microns. This value is needed by the program to convert into pixels positions. IMPORTANT: Leave this value at 1.0 to avoid unnecessary conversion between microns and pixels; so we define all our templates using pixel as basic unit (not microns). -
"gl_regions": Here we define regions in the template-image that contain growth lanes (see screenshot below). We can define multiple regions (e.g. for two-sided MMs). Each entry in"gl_regions"must define its own values for"horizontal_range","gl_spacing_vertical"and"fist_gl_position_from_top"even if they are identical:-
"horizontal_range": We assume the GLs to be oriented horizontally in the image (if not, image rotation needs to be set in preprocessing script accordingly). This range defines, where the GLs are located in the image (starting from the left side in pixels). You should specify a range larger than the actual GL length. In particular at the closed end of the GL you should leave a margin of >~20px to allow for correct U-Net processing. Also, you should include the bright fringes at the open end of the GL, so that they will be included in the ROI. These are ignored during processing with MoMA, but it is helpful to see what happens at the GL exit during curation of the tracking result. -
"gl_spacing_vertical": Vertical distance between the centers of two adjacent GLs in pixel. -
"fist_gl_position_from_top": The position fo the first GL as seen from the top of the image in pixel.
-
Notes regarding the creation of the template file and image:
- You can use ImageJ to create the template image and read the positions by moving the cursor to the corresponding position.
- You can use ImageJ to quickly create template-image crop and read the positions in pixel from that crop.
- Each entry in
"gl_regions"must define its own values for"horizontal_range","gl_spacing_vertical"and"first_gl_position_from_topeven"if they are identical.
A screenshot explaining the position parameters:
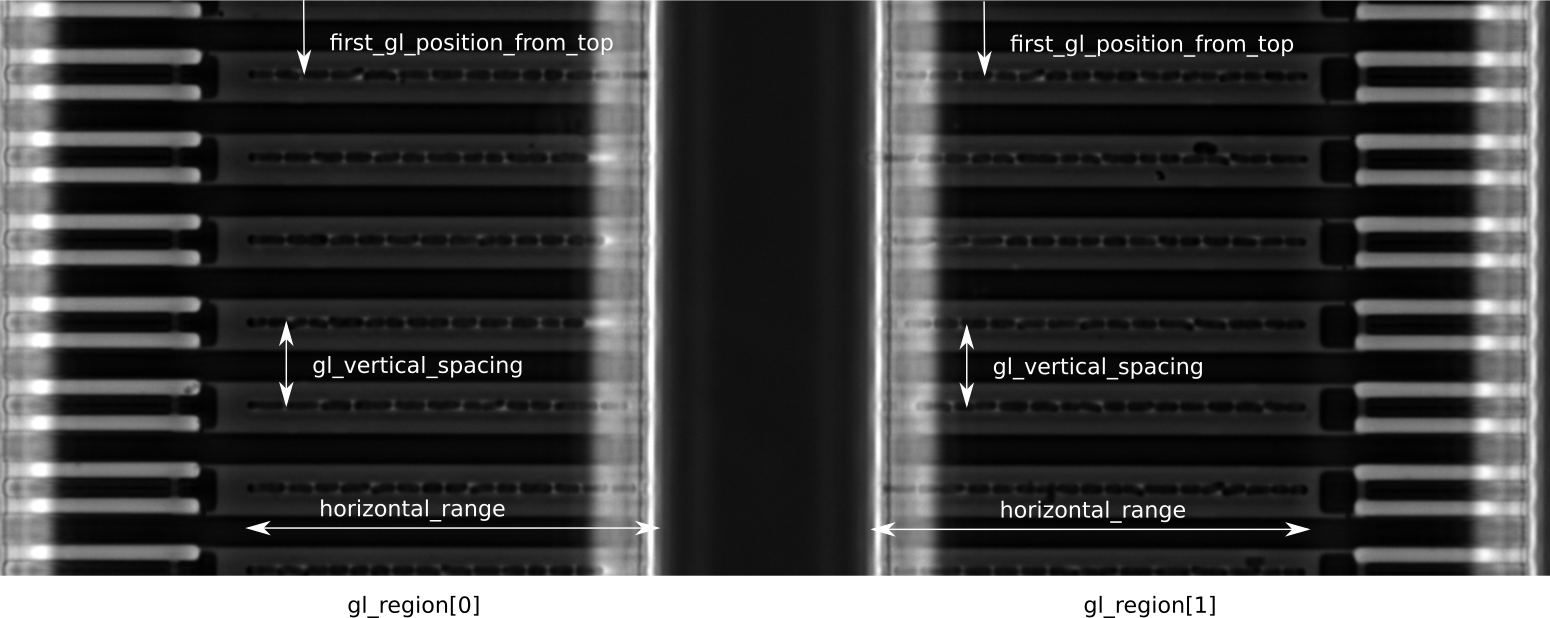
Things to consider for creating a GL template image:
- To create a template-image you can crop a region from any position of your experimental data. It should be as large as possible, but not be unique to the respective position (i.e. do not include any unique things such e.g. position-numbers).
- Make sure that the source-image is rotated so that the GLs are oriented horizontally in the image before making the crop. In ImageJ select Image -> Transformation -> Rotate ... to do so.
- From my testing, all GLs in the image should ideally be completely filled with cells. This suspect that this aids the template-matching, because full/dark GLs will contribute less in the calculation of the cross-correlation value than empty/bright GLs. This is preferable, since the appearance of GLs in the position-image will be different from position to position.
Todo: Link to MoMA introduction tutorial
To use a GL detection template we set the parameter
GL_DETECTION_TEMPLATE_PATH to point to the template configuration file
inside the SLURM script:
GL_DETECTION_TEMPLATE_PATH="/path/to/example_template_config.json"Note: * Without this parameter mmpreprocessingpy will still run and instead use a method of detecting GLs that does not require a template configuration.