-
Notifications
You must be signed in to change notification settings - Fork 0
Preprocessing Fluorescence Images
random edited this page May 29, 2017
·
2 revisions
This page describes how to analyze fluorescence image data in MoMA. If your data looks anything like this image, please read the following instructions.

In general the quick user guide is also applicable for fluorescence imaging. The additional steps listed below only apply to preprocessing, running MoMA should work the same way it does for phase contrast image data.
You can also try this using our example data.
- The first channel of the input file(s) has to contain the fluorescence image data. If your input data contains more channels and the fluorescence image is not placed first, please adjust the data. In Fiji this can be done using
Image→Stacks→Tools→Stack Splitter - In contrast to preprocessing phase contrast images, the image data has to be aligned straight (vertical growth lines). Slightly rotated input data will not be aligned during preprocessing and will therefore produce false results. You can rotate the data in Fiji using
Image→Transform→Rotate...
During growth channel separation two additional options are relevant.
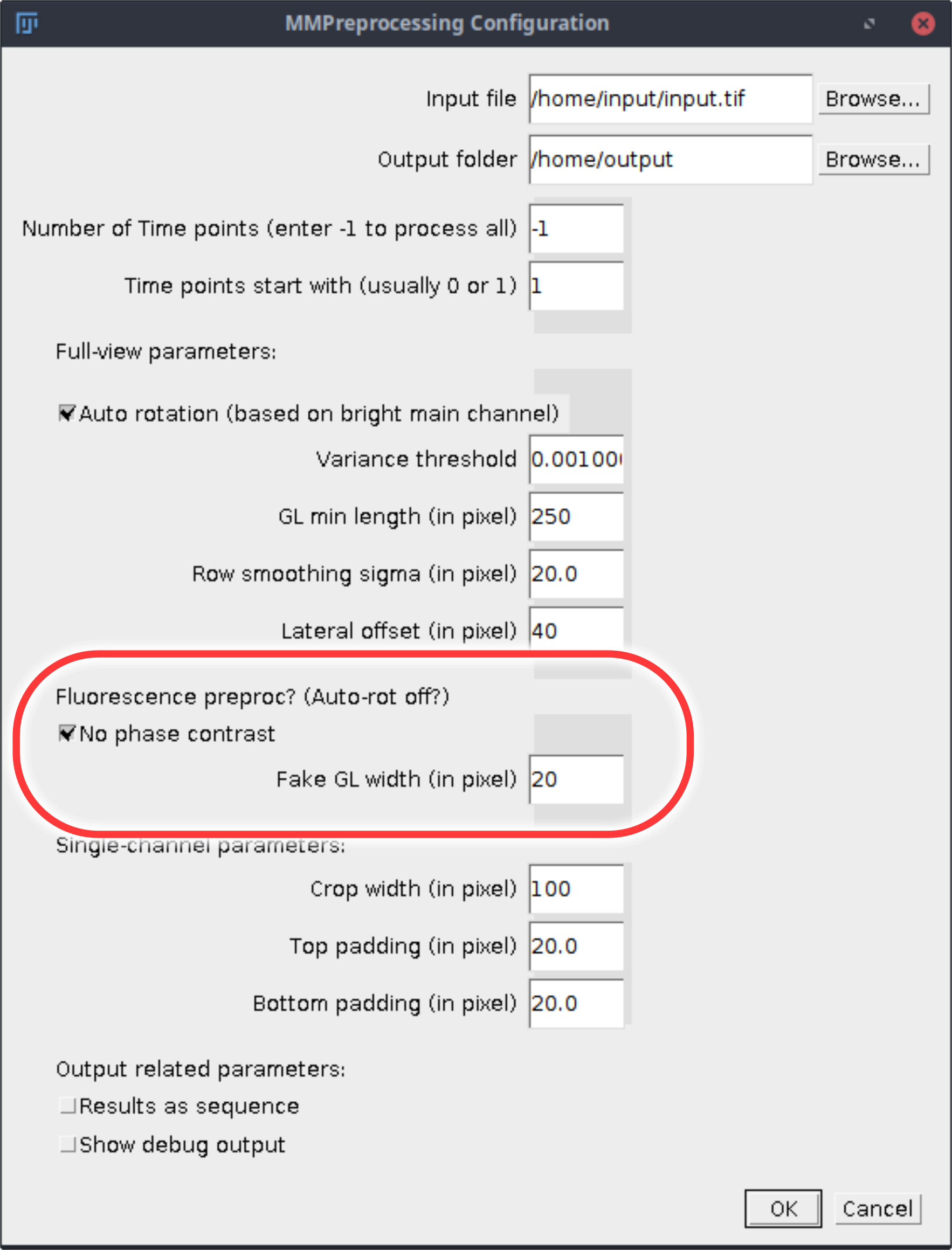
- Check
No phase contrast - Add the estimated width of the growth line to
Fake GL width (in pixel). To measure the width with Fiji use the line tool to draw a line horizontally through the growth line. Go toAnalyze→Set Scaleand copy the value shown inDistance in pixels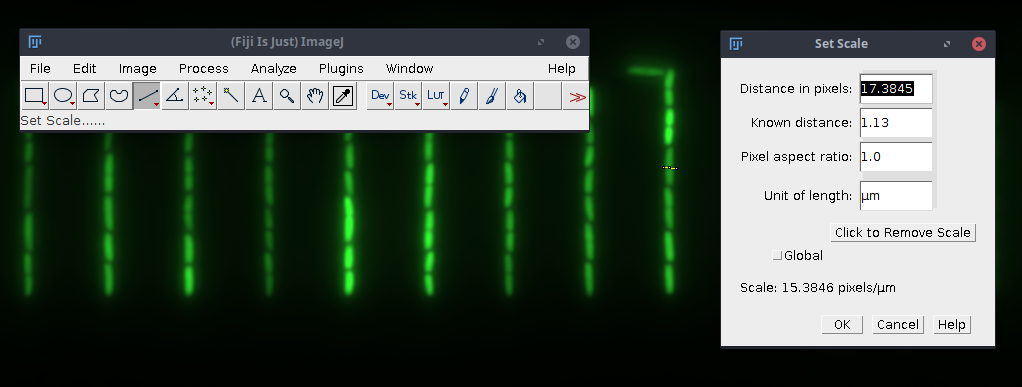
Please contact us if you are not able to analyze your fluorescence image data using MoMA.